Report Builders¶
Reports is an interactive graphical interface designed to facilitate data analysis and is based on the GOR language. The Reports tab opens by default when the Sequence Miner application is opened from the Clinical Sequence Analyzer (CSA) dashboard, displaying a grid which lists all available report modules.
To open any one of the modules in Sequence Miner, click on the module in the grid. The grid can be sorted according to the category/type of module.
Using Report Builders¶
The Reports tab contains a set of predefined report modules, also known as report builders, which can be used to parameterize a variety of different types of reports. There are over forty different modules currently available which target variant and gene annotation, Mendelian inheritance analysis, and cohort analysis use cases.
The Reports tab displays all available report builders as tiles. Clicking on a tile opens the selected report builder in a new tab, where input parameters can be edited to run the report.
Filtering and search¶
Report builders can be filtered by report type by clicking the arrow to open the drop-down menu. You can also search for individual report builders by entering keywords into the search bar, which searches report builder names, parameter names, and text descriptions.
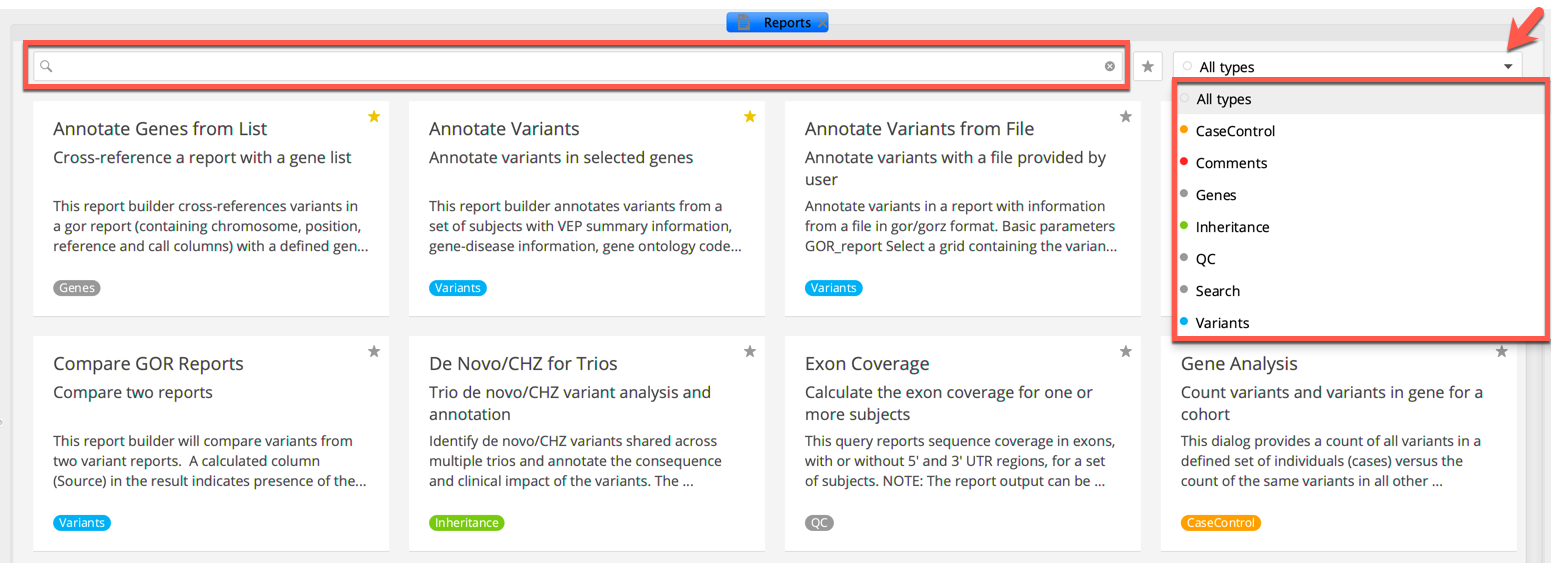
Filtering the Reports tab¶
Saving favorites¶
To save a report builder to your favorites, toggle the star icon in the upper right-hand corner of the selected report builder tile. To show only your favorite report builders, toggle the star icon next to the search bar.
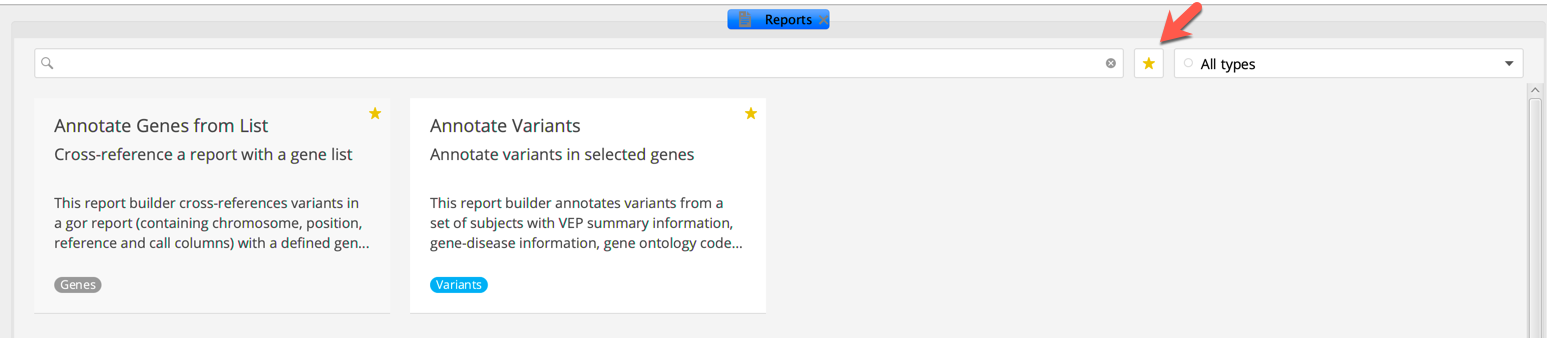
Clicking the star icon on a report adds it to your favourites¶
Reordering and undocking tabs¶
To reorder open tabs in the Sequence Miner window, click on a tab and drag it to a new location.
To undock a tab from the window, by right-click on the tab title and select Undock. To redock the tab into Sequence Miner, close the undocked window.
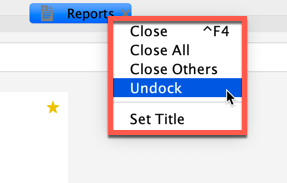
Right-click the tab title to undock¶
Renaming tabs¶
Tabs can be renamed by right-clicking the tab title and selecting Set Title.
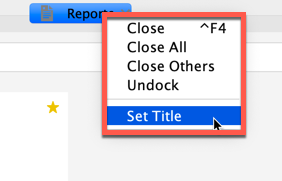
In the Set title dialog, enter a new title and click OK. Tab titles can include alphanumeric and underscore characters only.
Generating reports¶
When you have completed the form associated with any report, the Create Report button changes color from grey to light blue, indicating that you have entered sufficient input arguments to run the report. Click the button to generate the report.

Button changes color when sufficient data has been entered¶
Cancer Reports¶
The table below shows a list of the available report builders in Sequence Miner in the Cancer category with a short description of each. Note that Cancer modules are included only in selected installations of Sequence Miner and that your setup may be different.
Module Title |
Description |
|---|---|
Calculates and reports variant statistics for tumor samples |
|
Annotates and summarizes copy number variation (CNV) calls from the CNVkit caller for tumor samples |
|
Determine differentially expressed genes between two or more groups of samples |
|
Gene-aggregated variant analysis |
|
Provides the methylation status of samples at the individual probe level, gene level, or transcript level |
|
Provides expression sequencing data information for samples |
|
Provides miRNA expression sequencing data information for samples |
|
Count variants per VEP category |
|
Map samples across different data types available in the TCGA multi-omic dataset |
|
Counts the number of heterozygous, homozygous, and compound heterozygous variants in case and control subjects that pass filtering parameters |
|
Generate input table for statistical tools |
Case/Control Reports¶
The table below shows a list of the available report builders in Sequence Miner in the Case/Control category with a short description of each.
Module Title |
Description |
|---|---|
Generate a case-control grid from sets of subjects, for one or more phenotypes. |
|
dynamic_gwas |
GWAS analysis performed for the selected gene(s) or variant input, using genotypes from the selected freeze dataset. |
Count variants and variants in gene for a cohort. |
|
Gene-aggregated variant analysis. |
|
Count variants per VEP category. |
|
list_casecontrol_counts |
In a set of cases and controls, identify the count of ref, het, hom, and NA genotypes. |
list_subject_genotypes |
In a set of cases and controls, given a list of alleles (defined by variant or gene input), list the genotype for each individuals in each case/control category. |
Generate input table for statistical tools. |
|
Count the number of heterozygous, homozygous, and compound heterozygous variants in case and control subjects that pass filtering parameters. |
|
Evaluate the effect of variants on continuous or dichotomous traits using the sequence kernel association test (SKAT). |
|
skat_sequencing |
This report performs association tests between genotypes and a phenotype using the SKAT R package. |
Apply linear or logistic regression to quantify the relationship between specified variants and phenotypic data. |
|
single_point_regression_sequencing |
This report performs association tests between genotypes and phenotype(s) using the generalized linear model, which accepts continuous (e.g. glucose, IQ) as well as binary (e.g. case/control) variables. |
Single variant case-control association using Fisher Exact. |
|
Single variant case-control association using Fisher Exact for multiplicative (mm), recessive (rc), and dominant (dc) genetic models, using the Exome Aggregation Consortium (ExAC) database as controls. |
|
Count variants in a cohort. |
Gene Reports¶
The table below shows a list of the available report builders in Sequence Miner in the Gene category with a short description of each.
Module Title |
Description |
|---|---|
Cross-reference a report with a gene list |
|
Find carriers of variants in a genelist |
|
Quantify the association between a list of genes and categories in the Gene Ontology (GO) database |
|
Gene associated with Gene Ontology (GO) codes |
|
Maps/converts gene and protein IDs from reference source to all others and provide gene/protein names and IDs in a set of user-provided genomic regions |
|
Generate a report listing all paralogs of genes in a list |
|
Maps list of genes to corresponding pathway(s) |
|
Annotate variants with the disease(s) associated with genes and their paralogs |
|
Maps selected pathway(s) to a set of related genes in the same pathway(s) |
Inheritance Reports¶
The table below shows a list of the available report builders in Sequence Miner in the Inheritance category with a short description of each.
Module Title |
Description |
|---|---|
Preconception disease gene carrier analysis for parents |
|
Trio de novo variant analysis and annotation |
|
Single index case Mendelian disease analysis |
|
Zygosity analysis for cases versus controls |
Quality Control (QC)¶
The table below shows a list of the available report builders in Sequence Miner in the Quality Control (QC) category with a short description of each.
Module Title |
Description |
|---|---|
Calculate the exon coverage for one or more subjects using Ensembl transcripts |
|
Calculate the gene coverage for one or more subjects |
|
Calculate quality statistics for multiple subjects |
|
Check the sex of the samples |
|
Calculate the kinship of individuals |
|
Generate a variant summary for subject(s) |
Samples Reports¶
The table below shows a list of the available report builders in Sequence Miner in the Samples category with a short description of each.
Module Title |
Description |
|---|---|
Generate a case-control grid from sets of subjects, for one or more phenotypes. |
|
venn_diagram |
Venn comparison among up to ten grids containing a first column of sample identifiers. |
Search¶
The table below shows a list of the available report builders in Sequence Miner in the Search category with short description of each.
Module Title |
Description |
|---|---|
Identify subjects with information that matches the input keywords |
UKBB Reports¶
The table below shows a list of the available report builders in Sequence Miner in the UKBB category with a short description of each.
Module Title |
Description |
|---|---|
covariate_grid |
Produce covariate grids for use as input to subsequent analysis. |
phewas_lookup |
Finds records from genomic regions of interest with specified variant effect and p-value thresholds. |
uk_biobank_sets |
Search for relevant UKB fields and identify individuals with specific criteria for use as input to subsequent analysis. |
Variant Reports¶
The table below shows a list of the available report builders in Sequence Miner in the Variant category with a short description of each.
Module Title |
Description |
|---|---|
Annotate variants in selected genes |
|
Annotate variants with your own file |
|
Compare two reports |
|
Convert the genomic positions from one genome build to another |
|
Identify regions of loss of heterozygosity |
|
Count the variants within a gene list |
|
Return SNP genotypes for rsIDs |
|
Variant transcript effect |
|
Compare variant reports with Venn visualization |
Comments Reports¶
The table below shows a list of the available report builders in Sequence Miner in the Comments category with a short description of each.
Module Title
Category
Description
Internal annotations
Comments Reports
List variants with comments in local knowledgebase
Related comments
Comments Reports
Comments related to PN and gene