Sex check¶
The purpose of the Sex check query is identification of individuals with discordant biological sex assignment. This dialog checks whether the recorded biological sex and the inherent biological sex of study participants match.
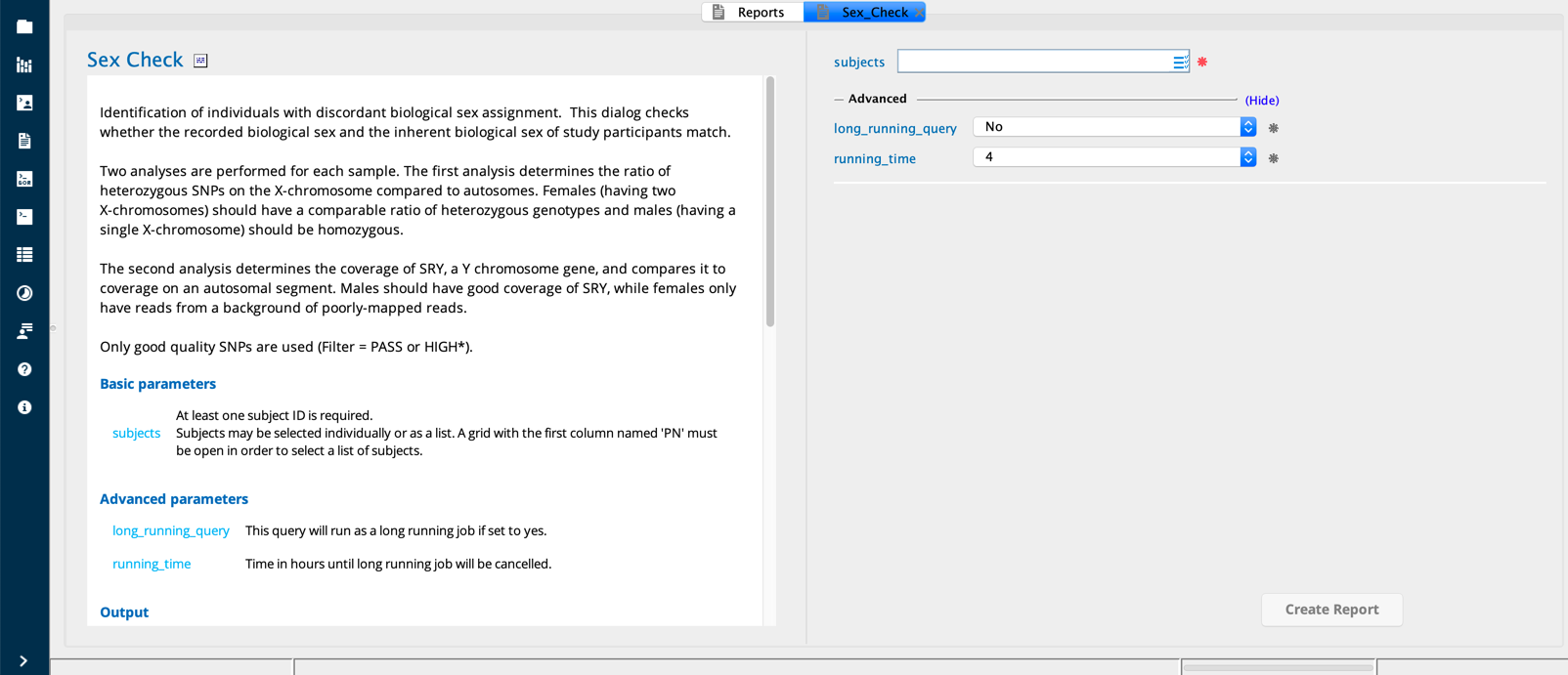
Sex Check module in Sequence Miner¶
Example use case¶
The user has a cohort of 200 subjects and wishes to confirm designated gender for all.
Description of the algorithm¶
Two analyses are performed for each sample:
The first analysis determines the fraction ratio (F_ratio) of heterozygous SNP calls for selected PNs on the X-chromosome and on chromosome 20 (the autosomal reference) with a minimum depth of 10 reads and good call quality calls. Females (having two X-chromosomes) should have a large set of heterozygous calls and males (having a single X-chromosome) should have few heterozygous calls.
Het_Fraction = het_calls / (het_calls + hom_calls)
F_Ratio = absolute(Het_Fraction20 - Het_FractionX) / Het_Fraction20
The second analysis determines the coverage of SRY, a Y chromosome gene, and compares it to coverage of a reference autosomal segment. Males should exhibit relatively high coverage of SRY, while females should only have a background of poorly-mapped reads. Coverage per bp is calculated across the SRY gene (ChrY:2,654,895-2,655,740) and a reference autosomal locus, Chr20:1,000,000-2,000,000. Coverage data comes from the
source/cov/segment_cov.gordfile.Coverage across SRY / Coverage across the reference region on chromosome 20
Interpreting the output¶
The F_ratio reflects the fraction ratio of heterozygous calls on the X chromosome to a reference autosome, chromosome 20. Samples with an F_ratio greater than 0.25 are annotated as “male”; samples with an F_ratio lower than 0.25 are annotated as “female”.
The SRYcovratio is the coverage of the SRY gene divided by the coverage of a 1Mb region on chromosome 20. The threshold is set to 0.2. Samples with a SRYcovratio greater than the threshold are annotated as “male”; samples with a SRYcovratio lower than the threshold are annotated as “female”.
Column descriptions¶
Group |
Column |
Description |
|---|---|---|
Basic |
Chrom |
Chromosome |
PN |
Subject ID |
|
Other columns |
bpStart |
|
bpStop |
||
F_ratio |
(value between 0 and 1) Ratio of heterozygous SNPs on the X-chromosome compared to heterozygous SNPs on autosomal chromosomes, calculated by: (ratio of het on X chromosome - ratio of het on chr20) / (ratio of het on chr20) |
|
GENDER |
(male/female) Sex of sample assigned by the system administrator at time of sample import |
|
SNPSEX |
(male/female) Sex determined by F_ratio. The threshold is set to 0.25 such that samples with an F_ratio greater than the 0.25 threshold are annotated as “male”; samples with an F_ratio lower than the 0.25 threshold are annotated as “female” |
|
SRYcovratio |
(value between 0 and 1) Coverage of the SRY gene divided by the coverage of a 1Mb region on chromosome 20 |
|
SRYSEX |
(male/female) Sex determined by the ratio of the SRY gene coverage versus the coverage of a portion of chromosome 20. The threshold is set to 0.2 such that samples with a SRYcovratio greater than the 0.2 threshold are annotated as “male”; samples with a SRYcovratio lower than the 0.2 threshold are annotated as “female” |
|
status |
|
Perspective views¶
Perspectives subtabs focus on subsets of the columns in the Default view.
Perspective |
Description |
|---|---|
Default view |
|
Sex Incorrect |