Query Editor¶
The Query Editor in Sequence Miner is a tool for writing and executing GOR queries on your genomic data.
To open the Query Editor, click the Data Query icon in the toolbar on the left-hand side of the Sequence Miner window. The editor opens in a new tab, which can be undocked by right-clicking the tab title and selecting Undock in the context menu.
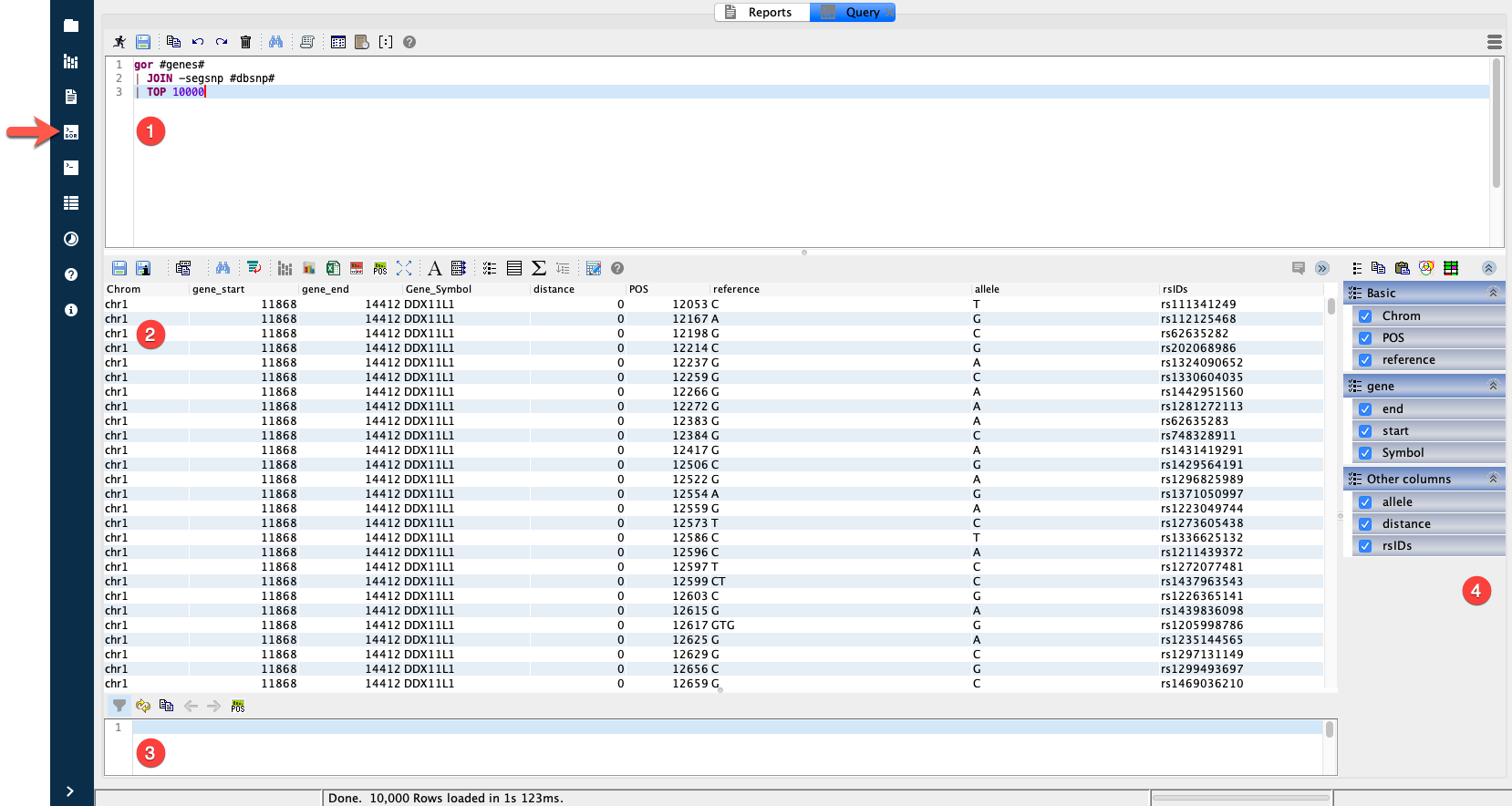
The Query Editor in Sequence Miner¶
The Query Editor has four main panels (numbered in the figure above):
Query input
Query output
Filtering
Column selector
Query input¶
The query input field is a syntax-aware tool for the GOR Query Language, a powerful tool for mining genomic-ordered data. The GOR Query Language is used to perform a wide variety of queries which underpin all other areas of Clinical Sequence Analyzer and Sequence Miner. For example, the Query Editor can be used to check the validity of the query behind a particular report or study type. The query input field also features code highlighting to aid in writing GOR queries.
Note
A full discussion of the GOR query language is outside of the scope of this document. For a full reference on writing GOR queries, please refer to the GOR Query Language manual, which can be accessed from the toolbar or by clicking the menu icon in the top right-hand corner.
The toolbar above the query input field provides access to a number of tasks that can be performed while working with queries. These are numbered in the image below and explained in the table that follows.

# |
Description |
Shortcut (Windows) |
Shortcut (Mac) |
|---|---|---|---|
1 |
Run the query |
CTRL+Enter |
⌘+Enter |
2 |
Save the query to a file |
CTRL+S |
⌘+S |
3 |
Copy all text from the editor to the clipboard |
||
4 |
Paste all text from the clipboard to the editor at the current location |
CTRL+V |
⌘+V |
5 |
Undo last change in the editor |
CTRL+Z |
⌘+Z |
6 |
Redo last undo in the editor |
CTRL+Y |
⌘+Y |
7 |
Delete the current line in the query editor |
CTRL+D |
⌘+D |
8 |
Search for text in the query editor |
CTRL+F |
⌘+F |
9 |
Format the query (puts each pipe command on a separate line) |
||
10 |
Display a list of available aliases |
F10 |
F10 |
11 |
Automatically complete GOR keywords (e.g., typing “GO” and using the keyboard shortcut opens a pop-up menu to select a matching GOR command) |
CTRL+Space |
⌘+Space |
12 |
Select a virtual relation for view |
||
13 |
Open the GOR Query Language help |
F1 |
F1 |
In addition, the following keyboard shortcuts can be used:
Description |
Shortcut (Windows) |
Shortcut (Mac) |
|---|---|---|
Comment out the selected code |
CTRL+/ |
⌘+/ |
Delete last word |
CTRL+Backspace |
⌘+Backspace |
Block comment: Select a comment block and press the shortcut keys |
CTRL+ALT+/ |
⌘+Option+/ |
Note
The keyboard shortcut for “Comment out the selected code” - CTRL+/ on Windows or ⌘+/ on Mac - may not work on computers with non-US keyboard layouts.
Aliases¶
GOR queries written in Sequence Miner frequently use aliases to refer to reference data files within GOR queries. To open a list of available aliases, click the Display alias list icon in the query editor toolbar.
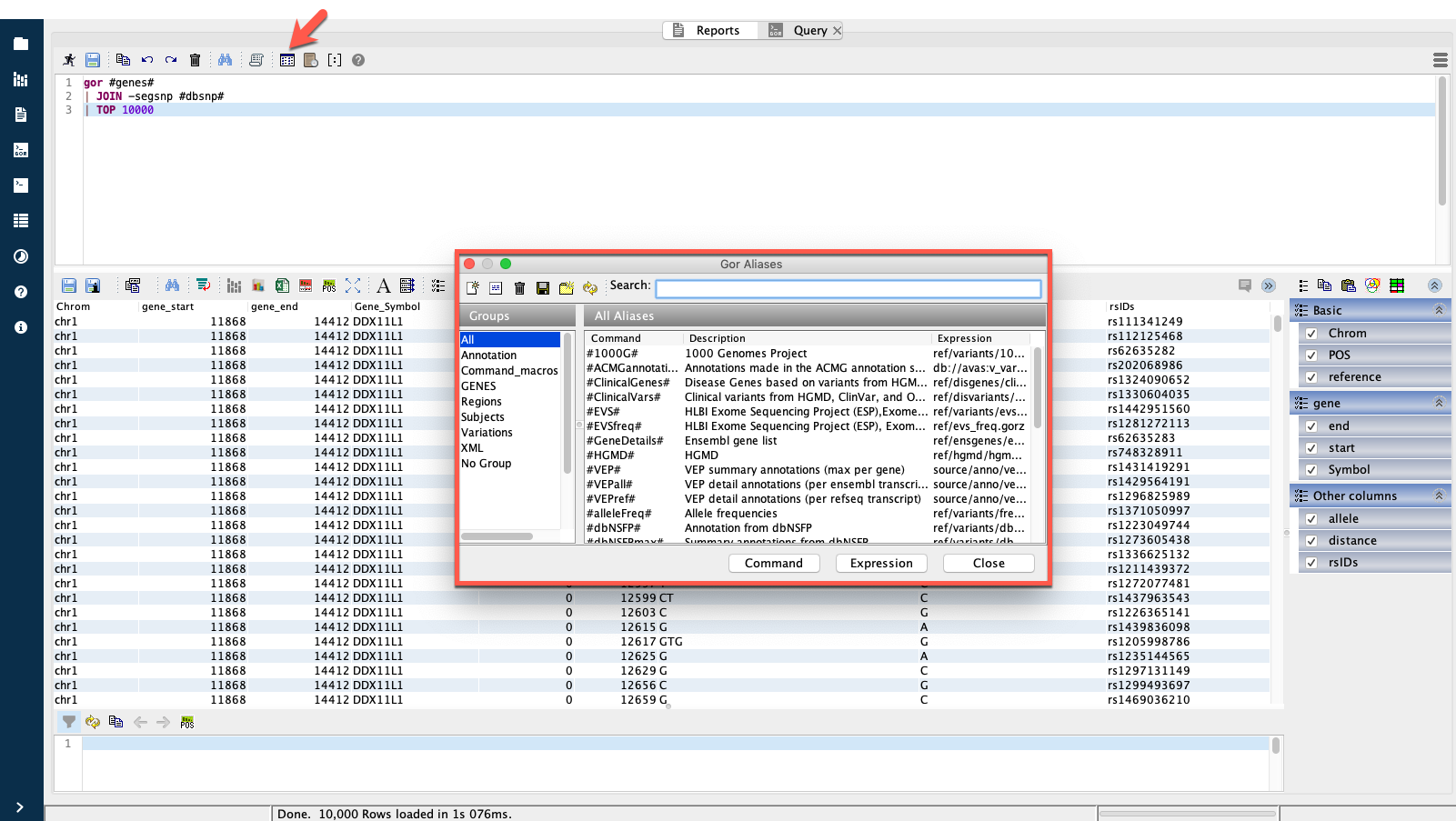
The full path to the files referred to in any alias can be expanded by selecting Replace Aliases from the hamburger menu.
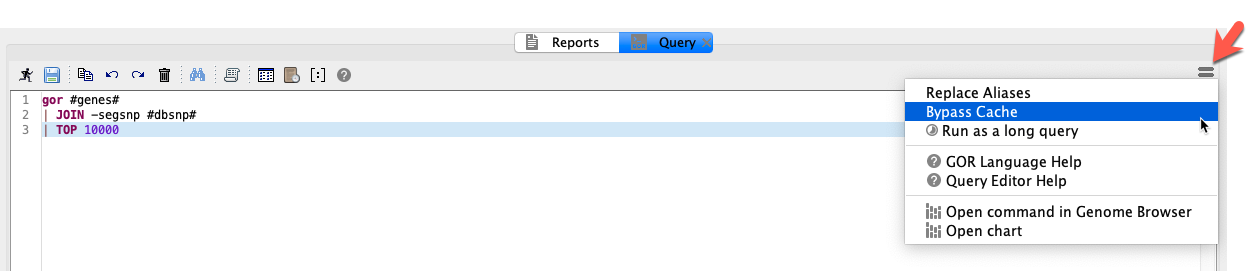
Cached queries¶
When a GOR query is run, the result of the query is cached for a period of time to speed up performance. To bypass the cache, click the menu icon in the top right-hand corner and select Bypass Cache.
Query output¶
When a query is run, the result is displayed in a table, called a grid, in the query output panel.
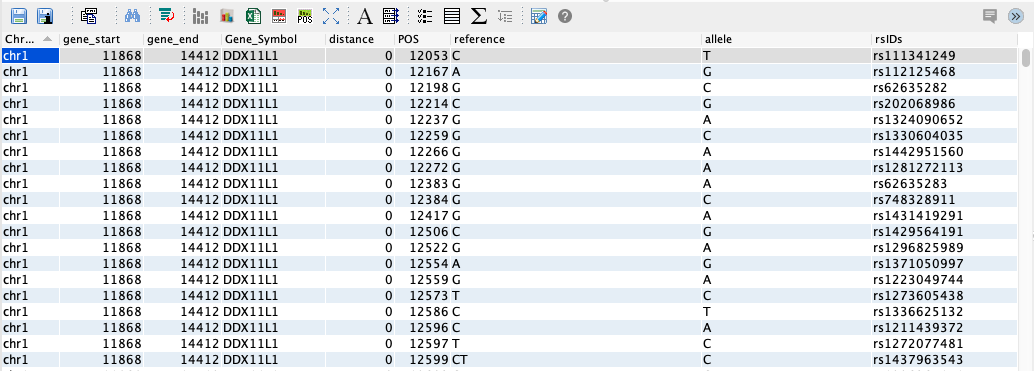
The toolbar above the query output field provides access to a number of tasks that can be performed on the output. These are numbered in the image below and explained in the table that follows.
# |
Description |
|---|---|
1 |
Save table to a file |
2 |
Save table to a file together with an info file |
3 |
Copy all selected cells with header |
4 |
Search table and select rows where found |
5 |
Reset table sort |
6 |
Show tracks in Genome Browser for selected rows |
7 |
Open chart |
8 |
Open in Excel |
9 |
Open track in Genome Browser |
10 |
Synchronize BAM file in Genome Browser |
11 |
Synchronize (right-click for details) |
12 |
Aggregate column values |
13 |
Add calculated column |
14 |
Show all columns |
15 |
Switch to record view |
16 |
Group |
17 |
Expand selected groups |
18 |
Edit table data |
19 |
Open Sequence Miner help |
For more detail about each function, see Navigating the output.