Genome Browser¶
The Genome Browser is a powerful analytical and diagnostic tool for finding variants in a subject or patient’s genome. The raw genomic data is presented alongside reference and annotation data to provide the user with a bird eye’s view of the sample data. Combined use of the Genome Browser and the Query Editor allows the researcher or clinician to drill down into the data and prepare detailed reports for a variety of use cases.
This section describes how to use the Genome Browser to work with your sample data.
The individual panels can be expanded or collapsed completely to change the view of the Genome Browser. It is possible to have multiple Genome Browsers open concurrently and to sync data between the open tabs. If your workstation has multiple screens, the tabs can be undocked to have different views open across your monitors.
The image below shows the different panels of the Genome Browser.
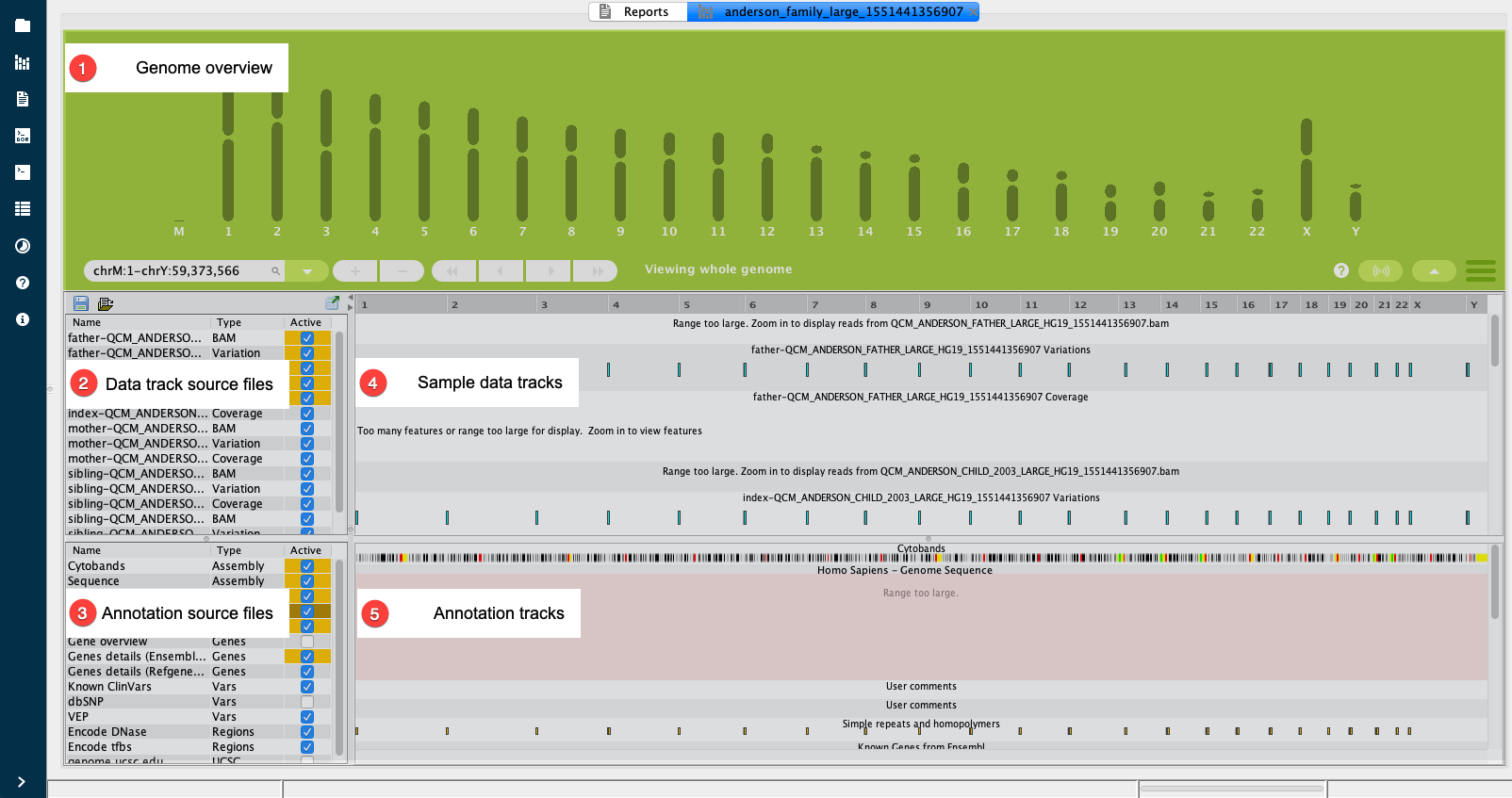
Genome Overview¶
The Genome Overview displays a visual representation of the human genome including chromosomes 1 to 22, both sex chromosomes, and a section at the beginning for mitochrondrial DNA. By clicking on any of these individual genome regions you can zoom in to the selected genomic range. When you click on a chromosome to zoom, the Genome Overview is minimized to provide a simple set of controls for selecting the range of data to display. You can expand the view again by toggling the arrow on the right-hand side.

Minimized Genome Overview showing only chr13¶
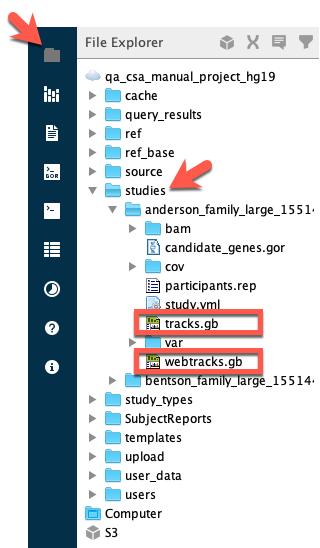
Genome Browser links in Sequence Miner have a .gb file extension. To quickly open the Genome Browser with all of the data and annotation tracks active, open the File Explorer and browse to your study. In the studies folder, there are two .gb files: tracks.gb and webtracks.gb. These files are created when you create a study in CSA. The file tracks.gb opens all of the data and annotation tracks.
Data tracks and annotation tracks are located below the Genome Overview, in four panels that contain a list of source files and a visual representation of each. To open a file in the visualization panel, select it in the corresponding list.
Data tracks¶
The following data tracks are available in Sequence Miner:
.bam- BAM files containing the output of secondary analysis in sequencing
.vcf- VCF files containing information about SNPs and small Indels
.cov- Coverage files
The way your data looks in the visualization panels will depend on the range that you have set using the Genome Overview. In the image below, the range selected is of the whole chromosome 13.
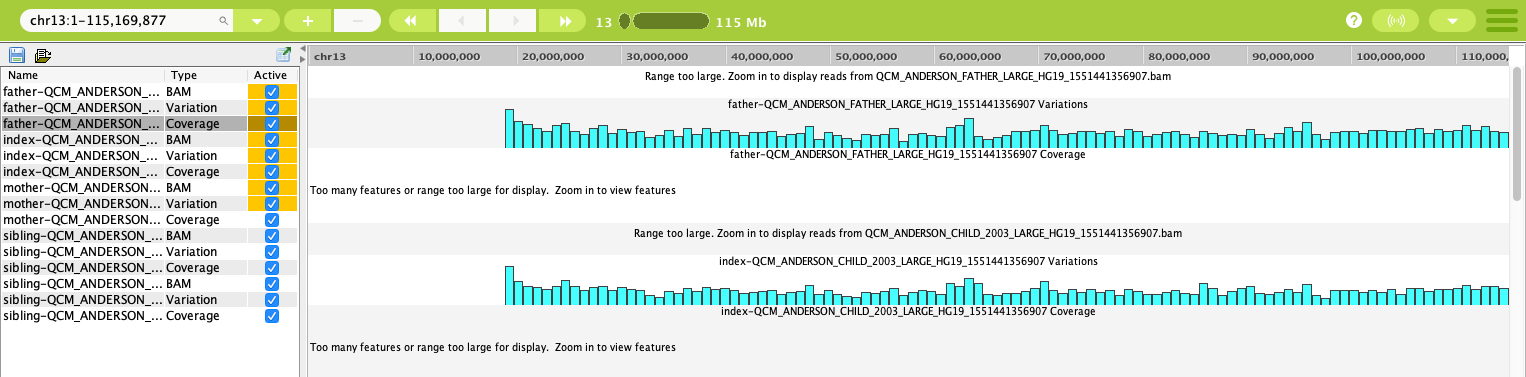
Data tracks in the Genome Browser with a large range¶
As you can see in the visualization panel, the range is too large to display all of the data correctly. Using the + and - buttons in the Genome Overview, you can zoom in and out on the tracks. You can also select a range directly in the header of the track, as shown below.
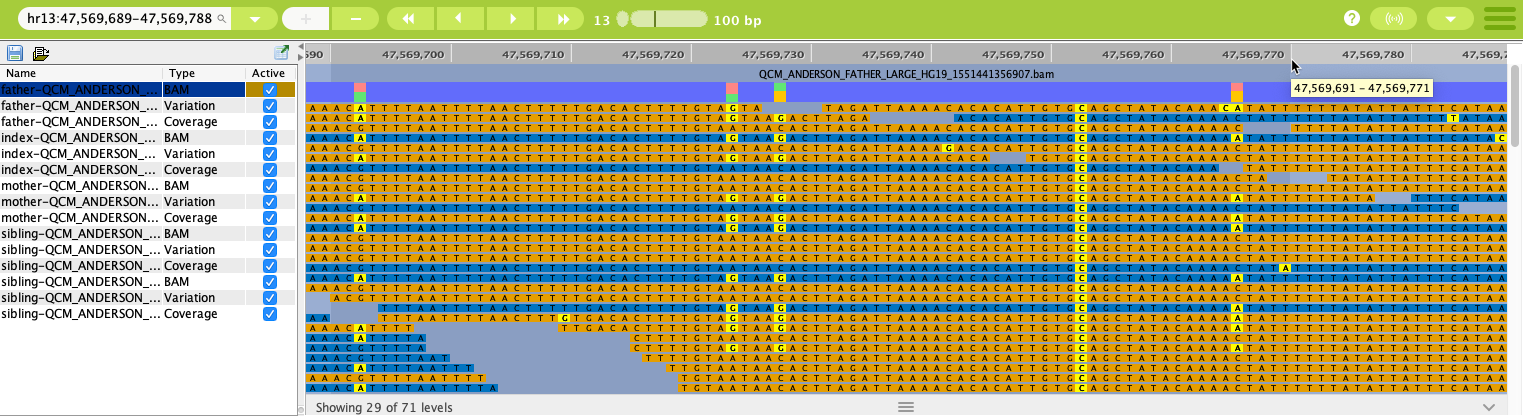
BAM data tracks in the Genome Browser with a shorter range shown¶
BAM tracks¶
Each line in the display above represents a single BAM track, and the tracks are color-coded according to various features of the tracks, with blue representing left reads and orange showing right reads. Deletions are represented in green and insertions in red. SNPs are represented by base positions with a yellow background. The colors of the tracks can be changed in Preferences.
Above the aligned stack of BAM tracks there is a section that indicates the percentage of each base call in that position for the sequenced read. To see the actual number of calls for each base at the position, you can enable tooltips on the BAM tracks using the context menu (right-click). If the top bar of the BAM track area is solid purple, then 85% or more of the calls were for the indicated base.
Right-clicking on an individual BAM track in the Genome Browser opens a context menu with several options.
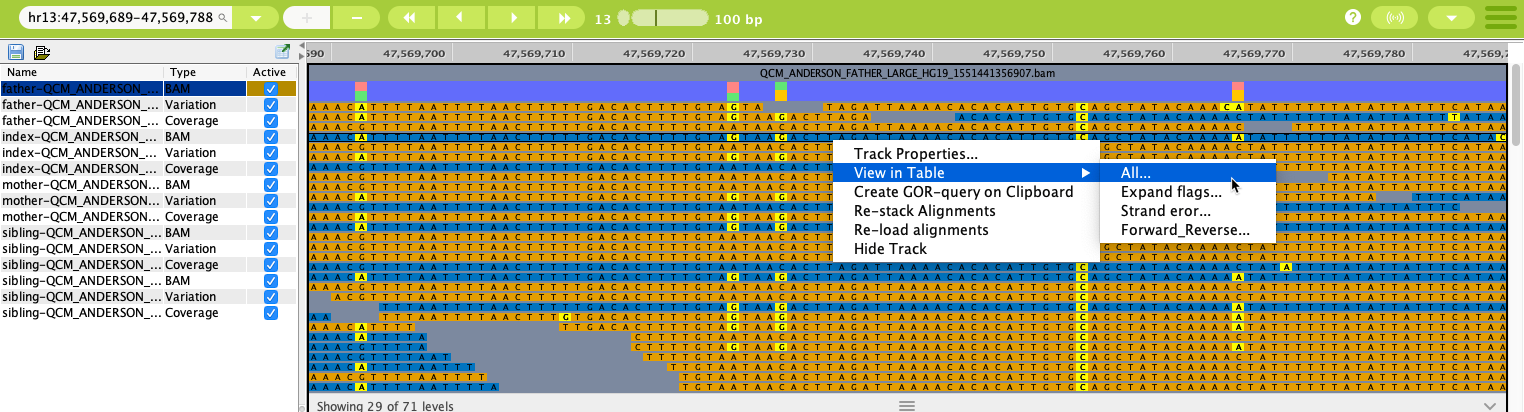
The right-click menu allows for deeper dives into the data¶
From the context menu, you can also open a Query Editor tab to show the GOR query for the displayed data.
See also:
Variation tracks¶
Variation tracks in the Genome Browser show the variation data for the individual sample that is selected and marked active in the data tracks. To display a variation track in the data track panel, the source file must be marked “Active” in the list on the left-hand side of the visualization panel.
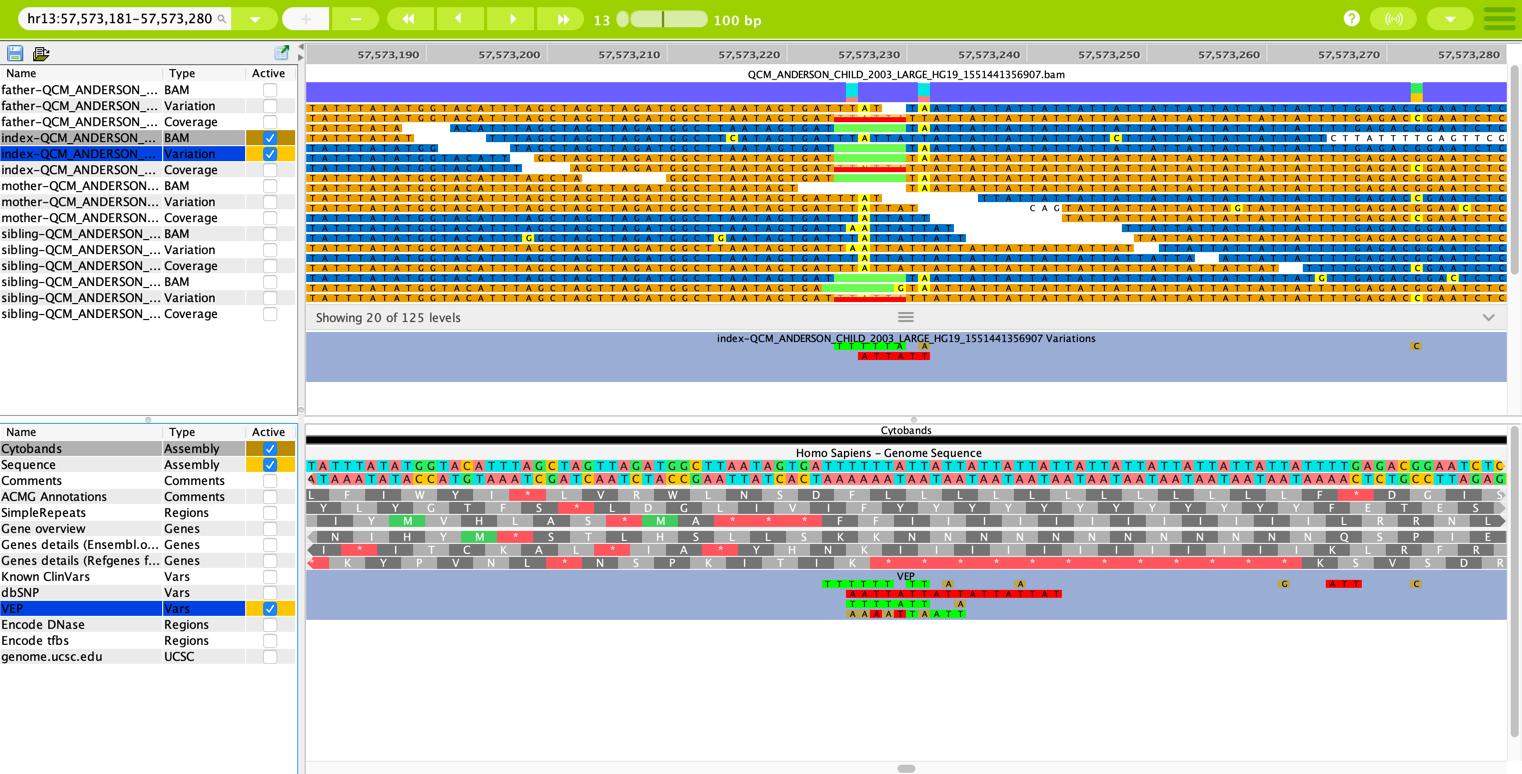
The variation tracks are shown below the BAM tracks and contain the variation data only¶
In the figure shown above, a single set of BAM and Variation tracks is displayed for the index case in this study. The BAM tracks show only the base value in the sequence where there is deviation from the reference genome sequence (which is shown in the annotation tracks below). The variations that are displayed here are two heterozygous deletions and two single-base SNPs, which can easily be compared to the VEP data in the annotation tracks in the panel below.
The variations are color-matched to the position where the variation occurs in the BAM track visualization.
Coverage tracks¶
Coverage tracks in the Genome Browser allow you to see the depth of the read at each genomic position in the sequence files. They can be represented graphically as bar graphs or scatter graphs. Hovering over an individual bar in the coverage graph opens a tooltip with information about the position range and the average depth of the coverage.
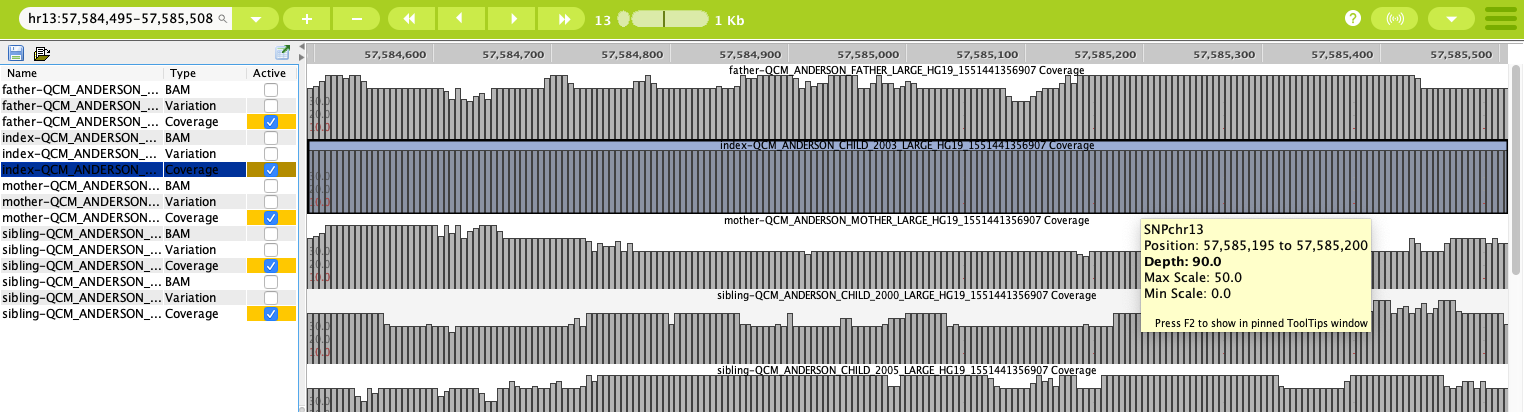
The coverage tracks can be set to display as bar graphs or scatter graphs to better visualize the information they contain¶
Right-clicking on the coverage graph opens a context menu where you select from several options, including opening the data in a table (showing the GOR query behind the graph) and reloading or hiding the track. Aggregated segments can also be created for files that have been loaded into Sequence Miner from your local computer, but not for remote sample files.
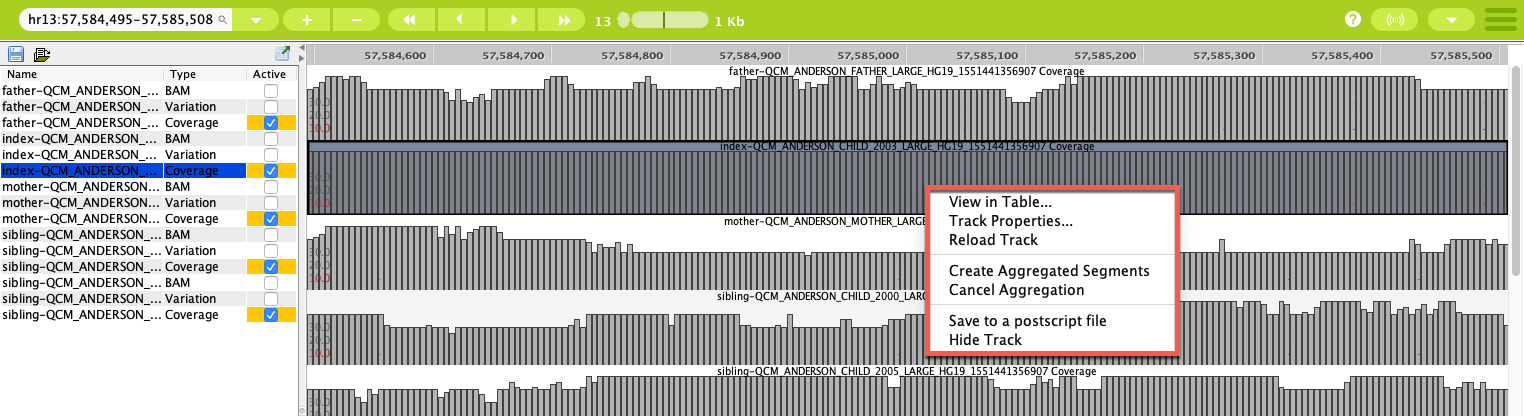
Right-click on the graph to open the context menu options¶
Selecting Track Properties in the context menu opens a Properties dialog where you can change display properties such as the color or style of the graphs.
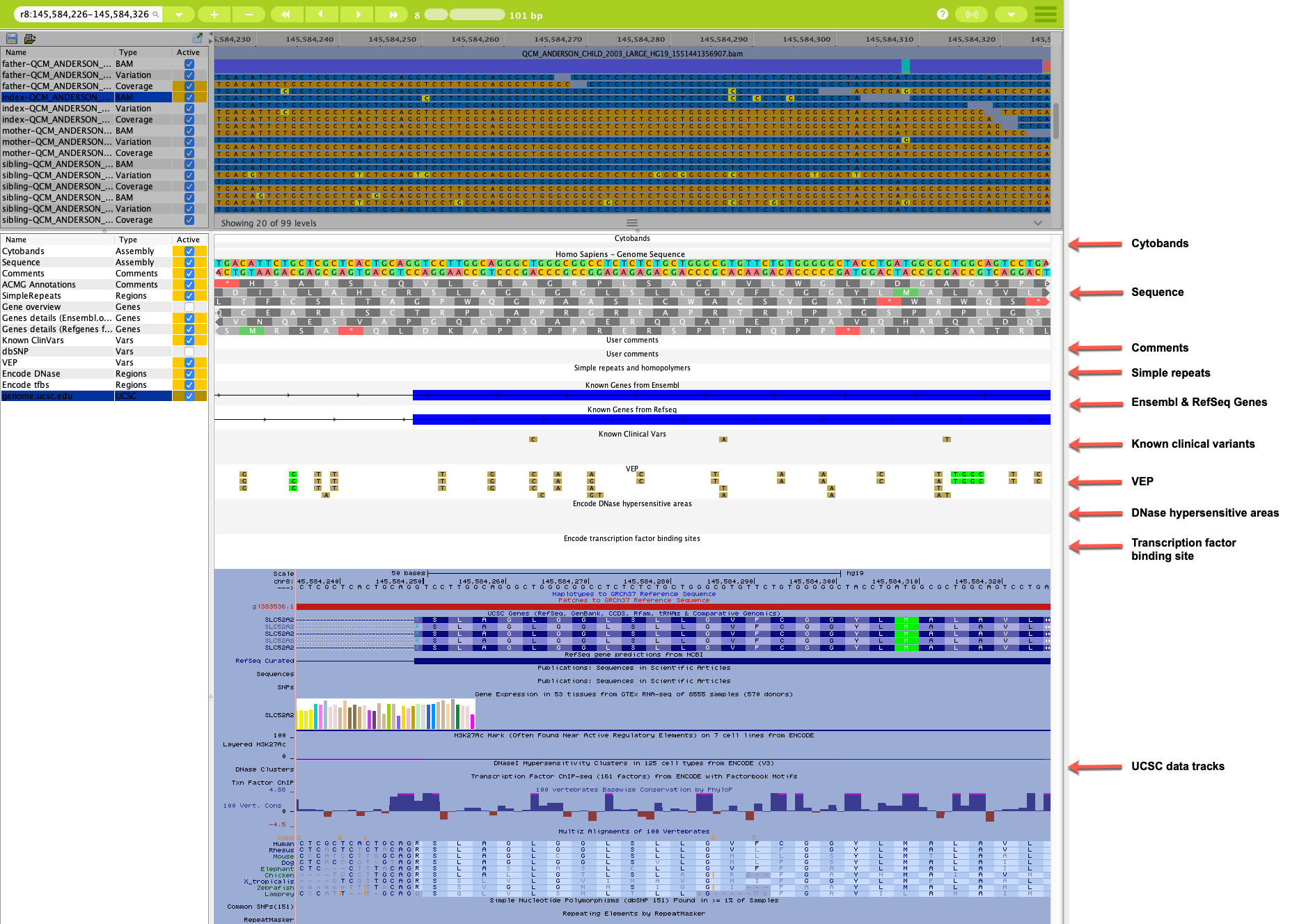
Annotation tracks¶
Fourteen default annotation tracks are listed in the Genome Browser. You can toggle the display of individual annotation tracks in the Active column by selecting the checkbox next to the track name. Tracks that are visible in the visualization panel are indicated by the orange background in the Active column.
The annotation tracks as they appear in the Genome Browser¶