BAM track preferences¶
To open the preferences for BAM tracks in the Genome Browser, right click anywhere in the BAM track panel and select Track Properties…
Editing view options¶
The Options tab of the Preferences for BAM Track is shown in the image below:
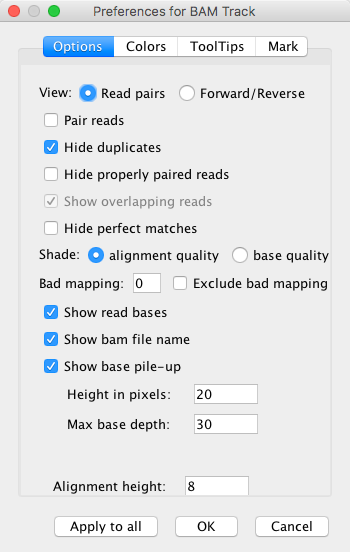
The Options tab in Preferences for BAM Track¶
In the Options tab, it is possible to change the visual representation of the BAM tracks in the Genome Browser. You can choose whether to view read pairs together or to show forward/reverse strands separately. Duplicate reads, properly paired reads, overlapping reads, and perfect matches can be shown or hidden from the view. Shading on the individual track colors can be also used to indicate either the alignment quality or base quality.
Other options are shown in the image above. The height of the tracks can also be edited.
Changing color preferences¶
The BAM tracks’ colors can be changed by right-clicking on the tracks, selecting Track Properties from the menu shown and selecting the Colors tab. Unclicking Color mate differently will display the contrast coloring of left and right reads.

Enabling tooltips on tracks¶
By default, no tooltips are shown for the BAM tracks. In the ToolTips tab in the Preferences for BAM Track, you can enable a list of the following tooltips, which will be shown on mouseover for each BAM track:
Mapping type
Read name
Alignment start
Alignment stop
Mate start
Alignment length
Inferred insert
CIGAR string
MD string
Mapping quality
Mate chromosome
Library
Marking individual bases¶
You can set the Genome Browser to mark bases according to a defined criteria in the Mark tab of the Preferences for BAM Track dialog.