CNV analyzer¶
The CNV Analyzer report builder annotates and summarizes copy number variation (CNV) calls from the CNVkit caller for tumor samples. This report builder provides an overview of CNVs discovered from tumor samples with annotation of known CNVs in similar locations previously reported in COSMIC.
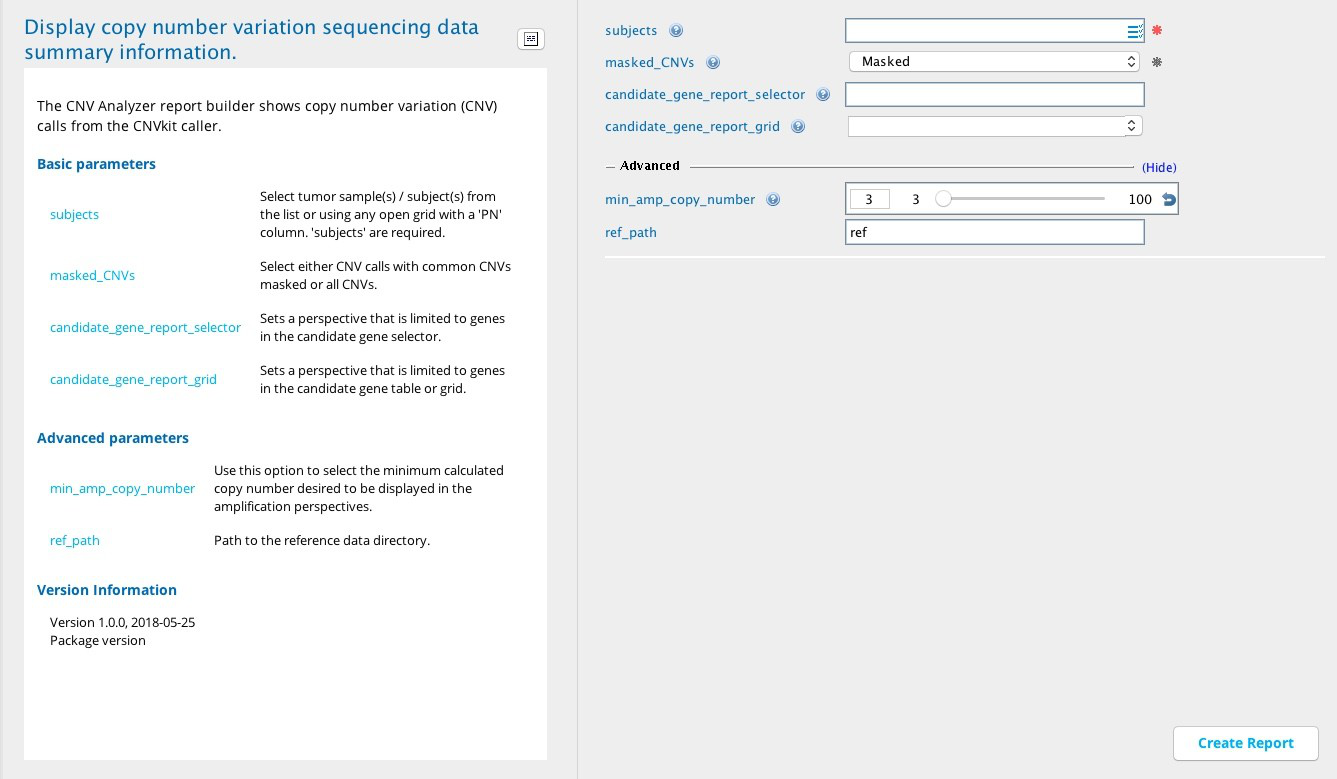
CNV Analyzer module in Sequence Miner¶
Example use case¶
The user is interested in identifying CNVs in specific tumor samples, genes involved in specific CNVs, and if any CNV in this location has been previously reported in COSMIC.
Description of the algorithm¶
CNVs from tumor samples selected from subjects are joined with Ensembl gene data to get the gene range for each CNV (if available), including total gene count, genes at the start position, and genes at the end position. If the CNV has the same gene range as CNVs previously identified in COSMIC CNV reference data, the COSMIC CNVs are listed in the COSMIC columns in the output, including CNV ID, tumor ID, primary site, etc. as annotation.
Interpreting the output¶
Output columns can be filtered by selecting one of the following Perspective views (see also Column descriptions):
Default view - Shows the CNVs identified from selected tumor samples which include the candidate genes provided in the candidate_gene_report selector or candidate_gene_report_grid input fields.
All Amplifications - Displays only CNVs with a copy number greater than the min_amp_copy_number.
All Loss and Deletions - Shows only CNVs with a copy number less than “2”.
Candidate Gene Amplification - Shows CNVs that include the candidate genes provided in the candidate_gene_report_selector or candidate_gene_report_grid fields and a copy number greater than the min_amp_copy_number.
Candidate Gene Deletions - Shows CNVs that include the genes provided in the candidate_gene_report selector or candidategene_report_grid fields and a copy number less than “2”.
Detailed CNV information such as Copy_number, mutation_type, Segment_Mean, Num_Probes, CNV_length, and CNV_size is displayed in the corresponding output columns.
The gene_symbol column lists the genes included in each CNV.
The Gene_range column shows the genes at the start position and end position of the CNV.
The PN and Flag column groups can provide information about the presence of other data types that are available for both the tumor sample and the corresponding normal sample, which can be useful for a multi-omic study.
Column descriptions¶
Group |
Column |
Description |
|---|---|---|
Basic |
Chrom, End |
CNV coordinates |
PN |
Sample/subject name selected individually or as part of a TCGA project as input from which the CNV is identified |
|
CNV |
gene_count |
Total number of genes included in CNV length |
length |
Length of the CNV in the chromosome in base pairs |
|
size |
Length of the CNV in the chromosome in KB or MB |
|
COSMIC |
CNV_ID |
Unique ID for a similar CNV (i.e., with the same gene range) reported in COSMIC |
gene_range |
The first and last gene included in CNV length in COSMIC |
|
ID_SAMPLE |
Sample ID of similar COSMIC CNV |
|
ID_STUDY |
Study ID of similar COSMIC CNV |
|
ID_TUMOR |
Tumor ID of similar COSMIC CNV |
|
MUT_TYPE |
Mutation type of similar CNVs: gain or loss |
|
Primary_histology |
Primary histology of similar CNV reported in COSMIC |
|
Primary_site |
Primary site of tumor of similar CNV reported in COSMIC |
|
SAMPLE_NAME |
Sample name of the similar CNV reported in COSMIC |
|
TOTAL_CN |
Copy number count of similar CNV reported in COSMIC |
|
Flag |
CNV_normal |
Logistic result whether the CNV normal sample is available |
CNV_tumor |
Logistic result whether the CNV tumor sample is available |
|
Methy_normal |
Logistic result whether the methylation normal sample is available |
|
Methy_tumor |
Logistic result whether the methylation tumor sample is available |
|
miRNA_norma |
Logistic result whether the miRNA normal sample is available |
|
miRNA_tumo |
Logistic result whether the miRNA tumor sample is available |
|
RNAseq_normal |
Logistic result whether the RNAseq normal sample is available |
|
RNASseq_tumor |
Logistic result wether the RNAseq tumor sample is available |
|
WES |
Logistic result whether the WES sample is available |
|
Gene |
range |
First gene and list gene included in CNV length |
symbol |
List of genes included in CNV length |
|
PN |
CNV_normal |
Paired normal sample ID for CNV |
Methyl_normal |
Paired normal sample ID for methylation |
|
Methyl_tumor |
Tumor sample ID for methylation |
|
miRNA_normal |
Paired normal sample ID for miRNA |
|
miRNA_tumor |
Tumor sample ID for miRNA |
|
RNASeq_normal |
Paired normal sample ID for RNAseq |
|
RNASeq_tumor |
Tumor sample ID for RNAseq |
|
WES |
Sample ID for WES |
|
Other columns |
Candidate_genes(option) |
List of candidate genes included in CNV length if they were specified as candidate_gene_report_selector or candidate_gene_report_grid input parameters |
Copy_number |
Total count of copies of the CNV |
|
Disease_type |
Disease type of the Subject in which the CNV was identified |
|
mutation_type |
Mutation type of the CNV: gain or loss |
|
Num_Probes |
Probe counts for the CNV |
|
Primary_site |
Primary site of the disease of the Subject |
|
Segment_Mean |
Log2 ratio of the tumor intensity to the normal intensity, which converts to an absolute copy number by (2^seg_mean)*2 |
|
startx |
Start position of the CNV |
|
SubjectID |
Patient ID |
|
TCGA_project |
Tumor type in TCGA project |
Perspective views¶
Perspectives subtabs focus on subsets of the columns in the Default view.
Perspective |
Description |
|---|---|
All Amplifications |
Shows all amplified CNVs (if CNV is located on the Y chromosome and the copy number is greater than “1”; if CNV is not located on the Y chromosome and the copy number is greater than the value of min_amp_copy_number, which is “3” by default) |
All Loss and Deletions |
Shows all deleted CNVs (if CNV is located on the Y chromosome and the copy number is equal to “0”; if CNV is not located on the Y chromosome and the copy number is less than “2”) |
Candidate Gene Amplifications |
Shows all amplified CNVs present in candidate genes (if selected in the input parameters) |
Candidate Gene Deletions |
Shows all deleted CNVs in candidate genes (if selected in the input parameters) |
Default view |
Shows all CNVs, including amplifications and deletions |