Sample map - TCGA¶
This report builder allows users to map samples across different data types available in the TCGA multi-omic dataset. TCGA data types include WES, CNV analysis, mRNA-Seq, miRNA-Seq and methylation analysis. Since samples of the same individual within each data type have different sample/PN IDs, a user may need to map the IDs across different data types. The report builder also allows users to retrieve sample/PN IDs of any data type using an individual’s SubjectID.
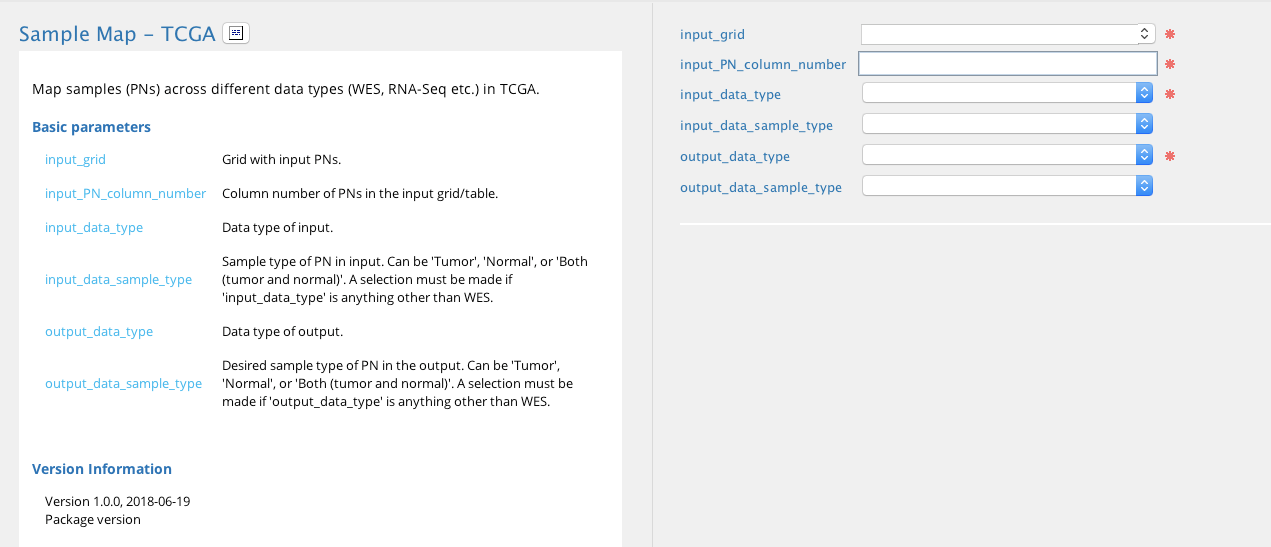
Sample map - TCGA module in Sequence Miner¶
Example use case¶
A user wants to obtain the differentially expressed genes between the High and Low TMB (tumor mutation burden) patients in the TCGA LUAD cohort. The user must first run the Tumor mutation burden report builder and filter the High and Low TMB patients. To be able to use these patients as an input for the Differential expression analysis using edgeR report builder, the user must obtain the corresponding mRNA IDs for these samples. The Sample map – TCGA report builder can be used to obtain this list of PNs/samples to use as cases and controls for differential expression analysis.
Description of the algorithm¶
The algorithm uses the SubjectReports/SamplePNs.rep file to map the input PNs of a given data type to the corresponding PNs of the desired data type.
Using the report builder¶
An input grid containing at least on column with PN/Sample/Subject IDs must be provided in the input_grid field. The data types of the input and desired output sample IDs must also be specified. For example, if the input grid contains CNV IDs and the desired output is the corresponding WES IDs (of the same individual) for that set of samples, CNV must be selected in the input_data_type field and WES must be selected in the output_data_type field.
Because the input grid may contain multiple columns, the column number that contains the input sample ID must be specified. For example, if the IDs of the input data type are in column 3 of the input grid, “3” must be entered in the input_column_number field.
In addition, the sample type (tumor, normal, or both) must also be specified for input or desired output data types mRNA, miRNA, CNV, and methylation. This is not required when WES or SubjectID is selected as the input or output data type.
Interpreting the output¶
The output displays the sample IDs of the desired data type. The output table/grid can be used as an input for other report builders to analyze the desired data type using the chosen report builder’s sample selector.
Column descriptions¶
Column |
Description |
|---|---|
PN |
Sample identifier of the output data type |
output_PN_type |
Output data and sample type |
input_PN |
Corresponding input PN that was supplied |