Gene/protein ID converter¶
This report builder maps/converts gene and protein IDs from one reference source to all others (e.g., Ensembl, RefSeq, HUGO, etc.). It also provides the gene/protein names and IDs in a set of user-provided genomic regions.
Example use case¶
Given a list of genes, the user wants to obtain the corresponding stable IDs from Enembl or Entrez sources.
To run the report builder, the ID/name type must first be selected in the input_id field (this is a required field).
For mapping/converting only a selected set of input gene/protein IDs or gene/protein IDs from particular genomic regions, the IDs or regions can be provided using either the input_file or input_grid fields.
Inputs include the following: BED file or genomic coordinates, gene symbol, HGNC symbol, Ensembl gene IDs, RefSeq IDs, Entrez IDs, HUGO gene symbols and Uniprot IDs. Note that the input_id field recognizes the first column of a grid unless “BED - custom regions” is selected, in which case the input file must be in BED format (chromosome, region starting position, region end position).
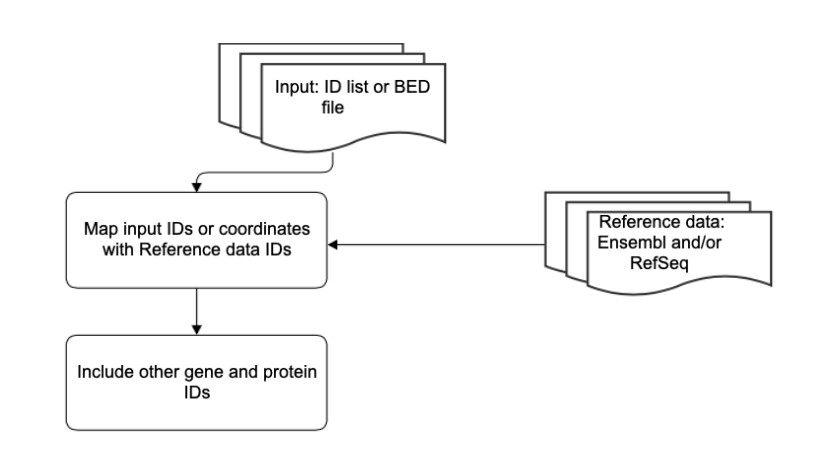
Description of the algorithm¶
Input IDs or coordinates are mapped with reference data sources including Ensembl, RefSeq, Entrez, HGNC and Uniprot to generate an output which includes the input ID and the corresponding IDs from the reference data files.
Interpreting the output¶
The output includes the input IDs or coordinates with the corresponding Ensembl or RefSeq gene symbols, Ensembl gene stable IDs, RefSeq IDs, Entrez Gene IDs, HGNC approved gene symbols, gene aliases, and Uniprot IDs.
Column descriptions¶
Group |
Column name |
Description |
|---|---|---|
Gene |
aliases |
The aliases of the given gene as defined by Ensembl or RefSeq |
End |
The end base pair position of the gene |
|
Start |
The start base pair position of the gene (zero based, i.e., the position of the base pair before the first base pair in the gene) |
|
Symbol |
The gene symbol from Ensembl or RefSeq (if symbols from both sources do not match, they will be separated in different rows) |
|
Other columns |
Chrom |
The chromosome where the gene (gene symbol) resides |
Coordinate_source |
Reference data source: Ensembl or RefSeq |
|
Ensembl_ID |
The Ensembl stable ID for the gene |
|
entrez_id |
The Entrez Gene ID |
|
HGNC_approved_symbol |
The HGNC (HUGO Gene Nomenclature Committee) symbol |
|
RefSeq_ID |
The RefSeq transcript (NM, NR, XM, XR, YP) or genomic (NC, NG) IDs |
|
Uniprot_ID |
The Uniprot protein ID |