Overlapping variants¶
The Overlapping variants query calculates how many variants in a variant GOR report reside within genes in a user-provided gene list in GOR format.
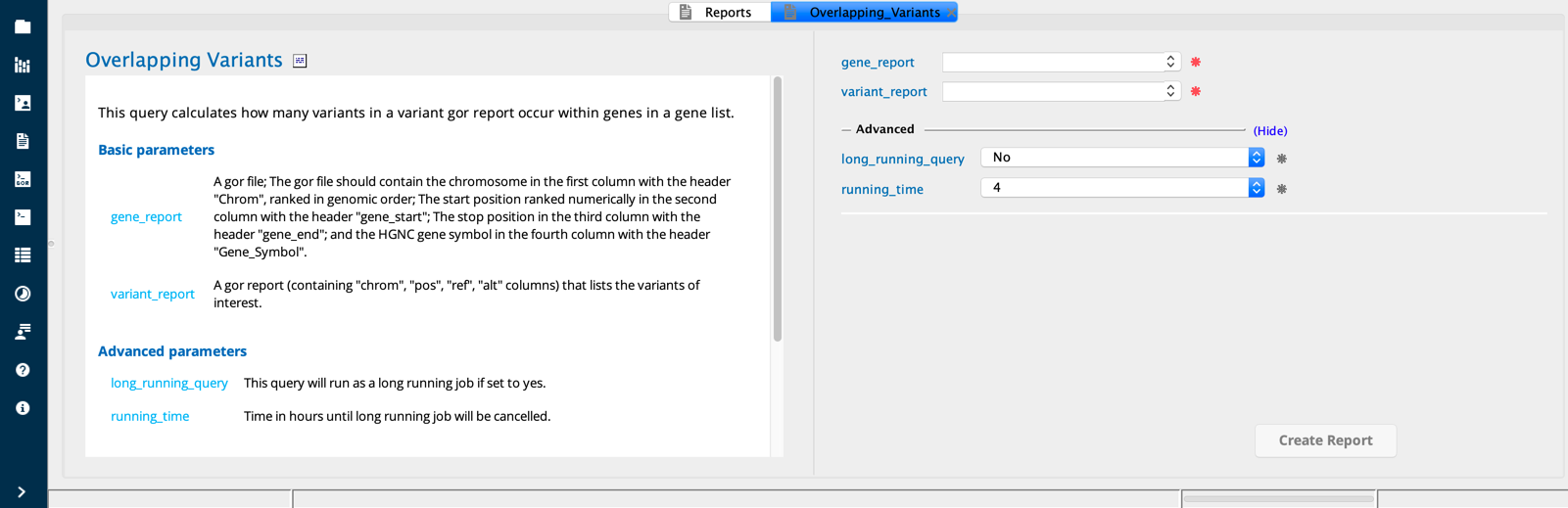
Overlapping Variants module in Sequence Miner¶
Example use case¶
The user wants to identify which variants in a variantreport (in GOR format) are present in a genereport (candidate gene list).
Description of the algorithm¶
For each gene in the gene list, this query joins a gene row in the genereport to the corresponding position of the variantreport if the gene segment overlaps the variant coordinate. The output includes an overlapVars column which displays a tally of variants mapped per gene.
Interpreting the output¶
The output is a GOR report with the columns described below. Annotation columns are drawn from the input genereport file.
Column descriptions¶
Group |
Column |
Description |
|---|---|---|
GENE |
end |
End coordinate |
start |
Start coordinate |
|
symbol |
Gene name |
|
Other columns |
Chrom |
The chromosome of the variant, represented as chr1, chr2, …, chr22, chrXY, chrX, chrY, chrM |
overlapVars |
The number of variants in a variant GOR report that occur within genes in the |