Gene association¶
The Gene association report (also known as the Gene Aggregate Variant Analysis (GAVA)) is based on a model in which disease-associated genes accumulate different variants across (unrelated) cases compared to controls. Rather than looking for over-representation of the same single gene variant in cases compared to controls, the accumulation of a variety of disease-associated variants can be detected as a difference in total variant load for a gene across cases as compared to controls.
The Gene Association or GAVA query first filters the input variants according to user-defined parameters including the following:
Variant scope (exonic only or whole genome)
Depth of sequencing coverage (1-50 reads)
VEP_consequence
Once variants have been filtered based on user-defined criteria, GAVA tests the hypothesis that disease related gene(s) has accumulated more variants across cases versus controls, or vice versa for protective variants. Each gene is scored independently so that multiple genes (e.g., in a common pathway or set of pathways) contributing to the phenotype may be identified.
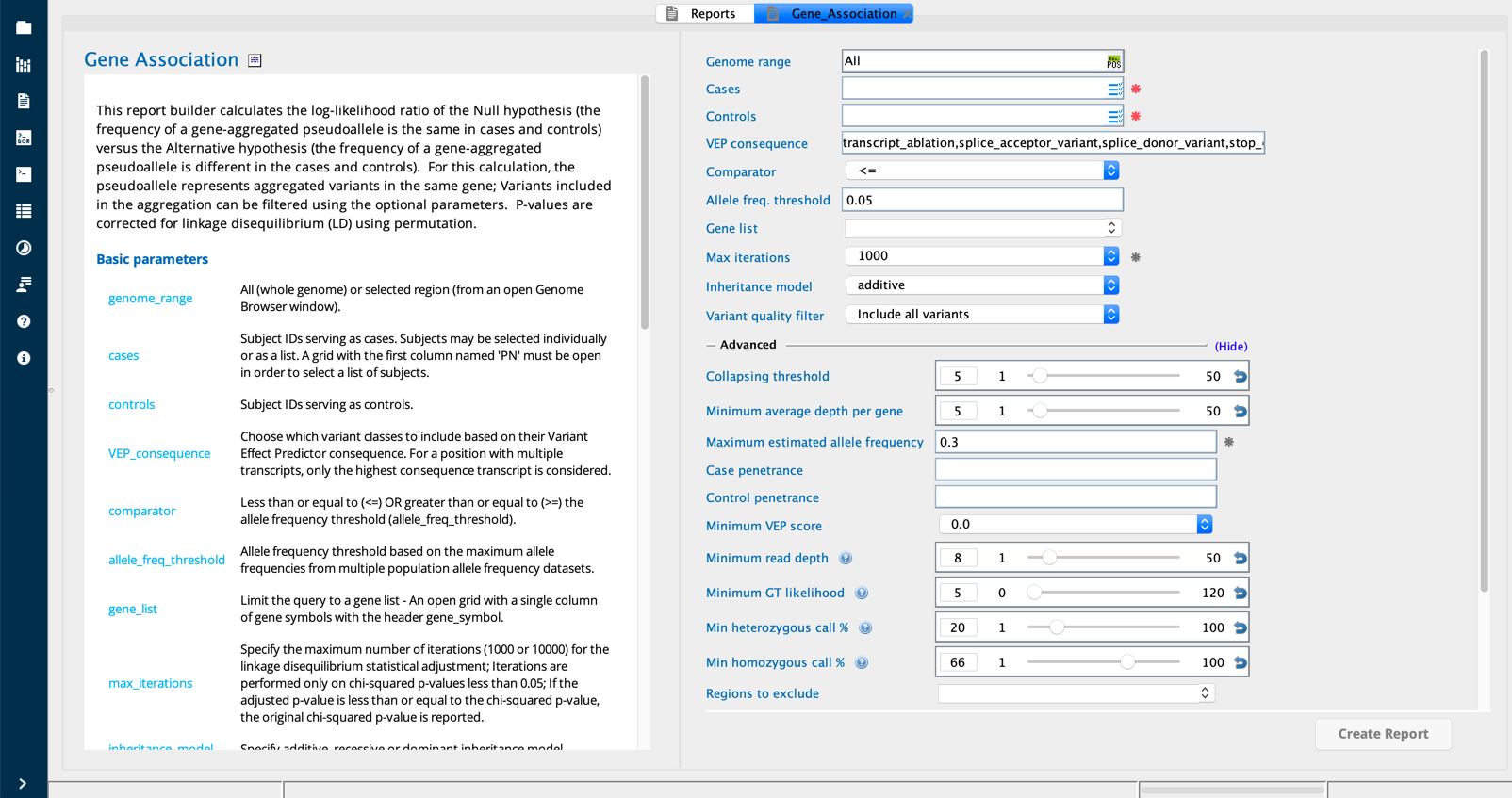
Gene Association module in Sequence Miner¶
Example use case¶
Given a whole exome sequence dataset for 1000 unrelated cases and 1000 controls, the goal is to identify genes carrying variants that contribute to the disease. The hypothesis is that disease-related genes have accumulated more variants across the cases compared to the controls.
Description of the algorithm¶
GAVA consolidates variants observed in each gene into an aggregate variant category based on frequency in Cases and Controls. GAVA then calculates the log likelihood ratio (LLR) of the aggregate variant category in Cases compared to Controls and returns the chi-square p-value (for the observed LLR value) and a permutation chi-square p-value. The permutation chi-square p-value corrects for linkage disequilibrium (a phenomenon in which variants are co-segregating). Because GAVA scores the LLR of the gene aggregate variant category load (across Cases versus Controls) rather than the LLR of each specific variant, GAVA does not rely on specific ancestral disease-causing variants, which is the model tested in a conventional GWAS study.
GAVA calculates the log odds ratio of the null hypothesis (Iikelihood of no difference) to the alternative hypothesis (likelihood of a difference):
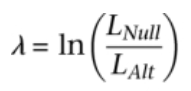
Interpreting the output¶
For each gene, a chi-square test score is calculated for the distribution of the aggregate variant load in Cases versus Controls. The associated chi-square p-value is reported in the output. In addition, a permutation p-value is calculated for genes with a chiPVal < 0.05. A minimum of 51 iterations are performed, which sets the threshold statistical significance level for 1000 iterations at a pValue of 0.05 (51/1000=0.051). The permutation is an iterative process of randomly resorting the sample IDs and recalculating the chi-square p-values of the aggregate variant. For each gene, the number of permutations performed is listed in the iterations column. The number of iterations is a count of the total permutations performed, of which a maximum of 51 can have a permuted p-value greater than the chi-square p-value for each gene. As a result, unless the maximum number of iterations were performed (e.g., 1000), the p-value/probability = 51 / #iterations. For example, if the number of iterations was 51, the permutation p-value will be 1.0 (51/51); if the number of iterations was 999, the permutation p-value will be 0.051 (51/999).
Use the chi-square p-value to rank genes by statistical significance of the difference in distribution of aggregate variant load between Cases and Control. The pVal column contains permutation pValues intended to account for linkage disequilibrium, meaning when genes variants are observed in the same gene because they are cosegregating rather than accumulating in the same gene independently. Once the user filters the output by some chiPVal threshold, the permutation pVal may then be used to filter out genes likely to have accumulated aggregate variants in Cases due to linkage disequilibrium.
Column descriptions¶
Group |
Column |
Description |
|---|---|---|
gene |
end |
|
start |
||
symbol |
||
Other columns |
chiPVal |
Chi-square p-value for the observed log likelihood ratio (LLR) of the aggregate variant category |
Chrom |
||
Iterations |
The number of iterations of randomizing (permutating) the sample IDs to calculate the permutation chi-square p-value. The permutation chi-square p-value is calculated only in cases where the chi-square p-value is < 0.05. The maximum number of iterations is 1000. The bailout is 51 iterations (no further iterations are performed when the number of times the permutation p-value > chi-square p-value reaches 51 prior to reaching the maximum set iterations). |
|
pVal |
The permutation p-value corrects for linkage disequilibrium (the situation in which variants in the same gene are cosegregating rather than arising independently) |
|
sum_LowCoverage |
For each gene, this value is the sum of cases and controls with average_depth (coverage) of less than 5 reads |