Variant association¶
The Variant association report builder performs a single variant case-control association analysis using Fisher’s exact test and an odds ratio calculation for three models:
The dominant model scores the difference in distribution of variant carriers (het or hom) among cases compared to controls.
The recessive model scores the difference in distribution of variant homozygotes among cases compared to controls.
The multiplicative model scores the difference in distribution of the variant alleles among cases compared to controls.
Variants detected in regions with coverage below a user-defined threshold or that fail a quality control filter are classified as “Unknown” and are therefore not used in the calculations.
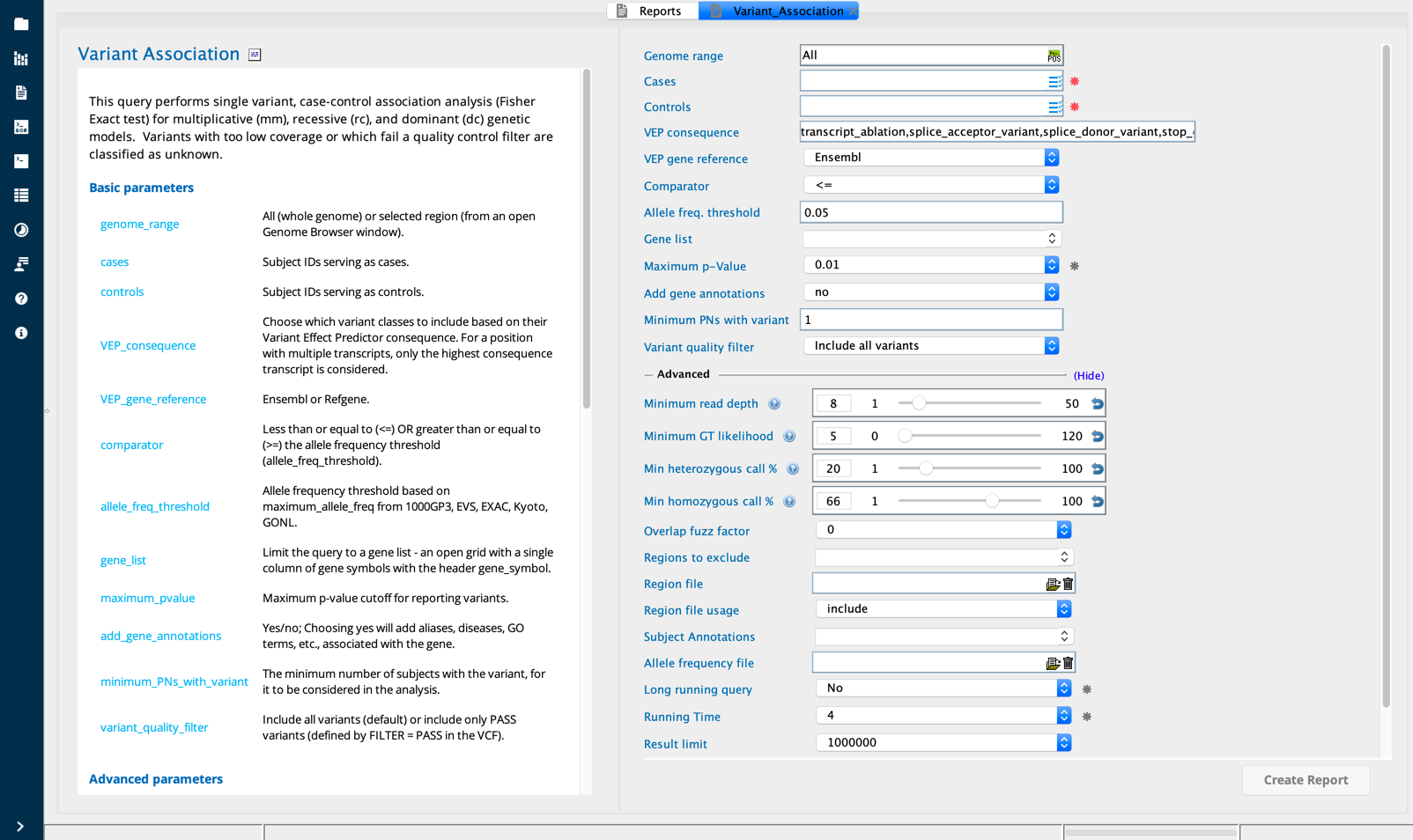
Variant Association module in Sequence Miner¶
Example use case¶
A user is interested in identifying variants with a statistically significant association with autism as compared to unaffected controls. Specifically, the user wishes to identify variants that are overrepresented in the patients versus the controls. Using a cohort of patients with autism (cases) and subjects without autism (controls), the user can perform a Fisher’s exact test and Odds Ratio calculation for each variant to measure the degree of association with the autism phenotype.
A Fisher’s exact test p-value < 0.05 indicates a statistically significant difference in the distribution of the variant in cases compared to controls. Therefore, the user might begin analyzing the results by sorting on the p-value columns for each inheritance model with the variants exhibiting the smallest p-values at the top of the list.
Description of the algorithm¶
For each variant identified in any Case or Control, an odds ratio and 2-tailed Fisher’s exact test p-value is returned for each of the 3 models (dominant, recessive and multiplicative).
For all samples from both case and control, a minimum coverage of, e.g., 8X (default) is required for the sample to be included in the calculation. The coverage information is included to eliminate potential false positives.
The Odds Ratio (OR) for each model is calculated as follows, using this general formula:
Odds Ratio = (A/B) / (C/D)
OR for the dominant inheritance model is calculated from the numbers of Cases and Controls carrying the variant allele.
Case_present is the number of cases carrying the variant (Cases homozygous and heterozygous for the variant allele).
Case_absent is the number of cases NOT carrying the variant (Cases homozygous for the reference allele).
Control_present is the number of controls carrying the variant (Controls homozygous and heterozygous for the variant allele).
Controls_absent is the number of controls NOT carrying the variant (Controls homozygous for the reference allele).
OR for the recessive inheritance model is calculated from the numbers of Cases and Controls homozygous for the variant.
Case_hom is the number of cases homozygous for variant.
Case_absent_hom is the number of cases NOT homozygous for the variant (Cases homozygous and heterozygous for the reference allele).
Control_hom is the number of controls homozygous for the variant.
Controls_absent_hom is the number of controls NOT homozygous for the variant (Controls homozygous and heterozygous for the reference allele).
OR for the multiplicative model is calculated from the numbers of variant and reference alleles in Cases and Controls.
Case_alleles is the number of variant alleles in cases.
Case_absent_alleles is the number of reference alleles in cases.
Control_alleles is the number of variant alleles in controls.
Controls_absent_alleles is the number of reference alleles in controls.
The 2-tailed Fisher’s exact test is performed for each model using the following input for a 2 X 2 contingency table:
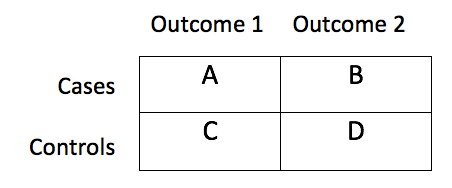
Interpreting the output¶
A 2-tailed Fisher’s exact test p-value is returned. A p-value < 0.05 indicates a statistically significant difference in the distribution of the number of heterozygous and homozygous cases compared to controls (dominant model), homozygous carriers compared to controls (recessive model) or variant alleles in cases compared to controls (multiplicative model).
An OR > 1 indicates a difference in the distribution of heterozygous carriers (dominant model), homozygous carriers (recessive model) or the variant allele (multiplicative model) in cases compared to controls.
Here is an example of input (counts) and output (OR and pVal):
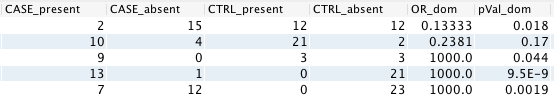
Dominant model¶
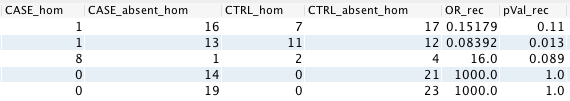
Recessive model¶
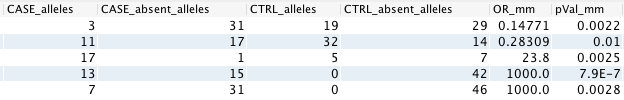
Multiplicative model¶
Column descriptions¶
Group |
Column |
Description |
|---|---|---|
Basic |
Call |
|
Chrom |
||
POS |
||
Reference |
||
CASE |
absent |
The number of cases in which the variant allele is absent |
absent_alleles |
The total number of reference alleles in cases |
|
absent_hom |
The number of cases that are not homozygous for the variant allele |
|
alleles |
The total number of variant alleles in cases |
|
het |
The number of cases that are heterozygous for the variant allele |
|
hom |
The number of cases that are homozygous for the variant allele |
|
present |
The number of cases in which the variant allele is present |
|
prop_alleles |
The proportion of variant alleles/total alleles with good coverage in the case group |
|
prop_het |
The proportion of individuals that are heterozygous for the variant allele/total number of individuals with good coverage in the case group |
|
prop_hom |
The proportion of individuals that are homozygous for the variant allele/total number of individuals with good coverage in the case group |
|
total_alleles |
The total number of alleles with good coverage in the case group |
|
total_PNs |
The total number of individuals with good coverage in the case group |
|
unknown |
The number of cases missing the variant allele and have low coverage (e.g., default is minimum 8X coverage) or a poor quality call at this position |
|
CTRL |
absent |
The number of controls in which the variant allele is absent |
absent_alleles |
The total number of reference alleles in controls |
|
absent_hom |
The number of controls that are not homozygous for the variant allele |
|
alleles |
The total number of variant alleles in controls |
|
het |
The number of controls that are heterozygous for the variant allele |
|
hom |
The number of controls that are homozygous for the variant allele |
|
present |
The number of controls in which the variant allele is present |
|
prop_alleles |
The proportion of variant alleles/total alleles with good coverage in the control group |
|
prop_het |
The proportion of individuals that are heterozygous for the variant allele/total number of individuals with good coverage in the control group |
|
prop_hom |
The proportion of individuals that are homozygous for the variant allele/total number of individuals with good coverage in the control group |
|
total_alleles |
The total number of alleles with good coverage in the control group |
|
total_PNs |
The total number of individuals with good coverage in the control group |
|
unknown |
The number of controls missing the variant allele and have low coverage (e.g., default is minimum 8X coverage) or a poor quality call at this position |
|
MAX |
AF |
Maximum reported allele frequency across the population surveys from 1000GP3, EVS, EXAC, Kyoto, GONL |
Impact |
Classification of the level of severity of the transcript consequence type assigned by VEP |
|
OR |
dom |
OR for a dominant inheritance models is calculated from the number of cases and controls carrying the variant allele |
mm |
OR for a multiplicative inheritance model is calculated from the number of variant and reference alleles in cases and controls |
|
rec |
OR for a recessive inheritance model is calculated from the number of homozygous genotypes (homozygous for the variant allele) in cases and controls |
|
pVal |
dom / rec / mm |
A p-value less than 0.05 (or 0.01) indicates a statistically significant difference |
Note on the Odds Ratio (OR)¶
The output will be an Odds Ratio (OR) for each variant for each of the three models.
OR > 1: The number of variant carriers, homozygotes or variant alleles among Cases is larger than the number among Controls. The higher the value, the more significant the difference.
OR < 1: The number of variant carriers, homozygotes or variant alleles among Cases is smaller than the number among Controls. The lower the value, the more significant the difference.
OR = 1: No difference in the distribution of variant carriers, homozygotes or variant alleles among Cases versus Controls.
OR = 1000: The number cases_absent or controls_present = 0.
The following are three exceptions to the standard OR calculation as defined above:
If any of the following values = 0: case_absent, case_absent_hom, case_absent_alleles, controls_present, controls_present_hom, or controls_present_alleles then the Odds Ratio = 1000. This number was chosen to represent complete association because if B or C is “0” in the Odds Ratio formula, an error is returned: OR = (A/B) / (C/D).
If case_present is 0, then the Odds Ratio = 0.
If any of the following values = 0: control_absent, control_absent_hom, or control_absent_alleles then the Odds Ratio = “NA”.
Perspective views¶
Perspectives subtabs focus on subsets of the columns in the Default view.
Perspective |
Description |
|---|---|
Default view |
|
Dominant_model |
|
Multiplicative_model |
|
Recessive_model |
Drill-in reports¶
Drill in |
Description |
|---|---|
VarCaseCtrlCarriers |
This drill-in report lists all carriers (from the cases and controls Sample ID lists) with the selected variants |
GeneCaseCtrlCarriers |
This drill-in report lists all carriers (from the cases and controls Sample ID lists) with variants in the selected genes |