Gene carrier report¶
The Gene carrier report builder finds carriers of variants in a user-designated genelist among subjects in a user-designated list. The user may limit the report to variants that meet VEP_consequence and quality thresholds.
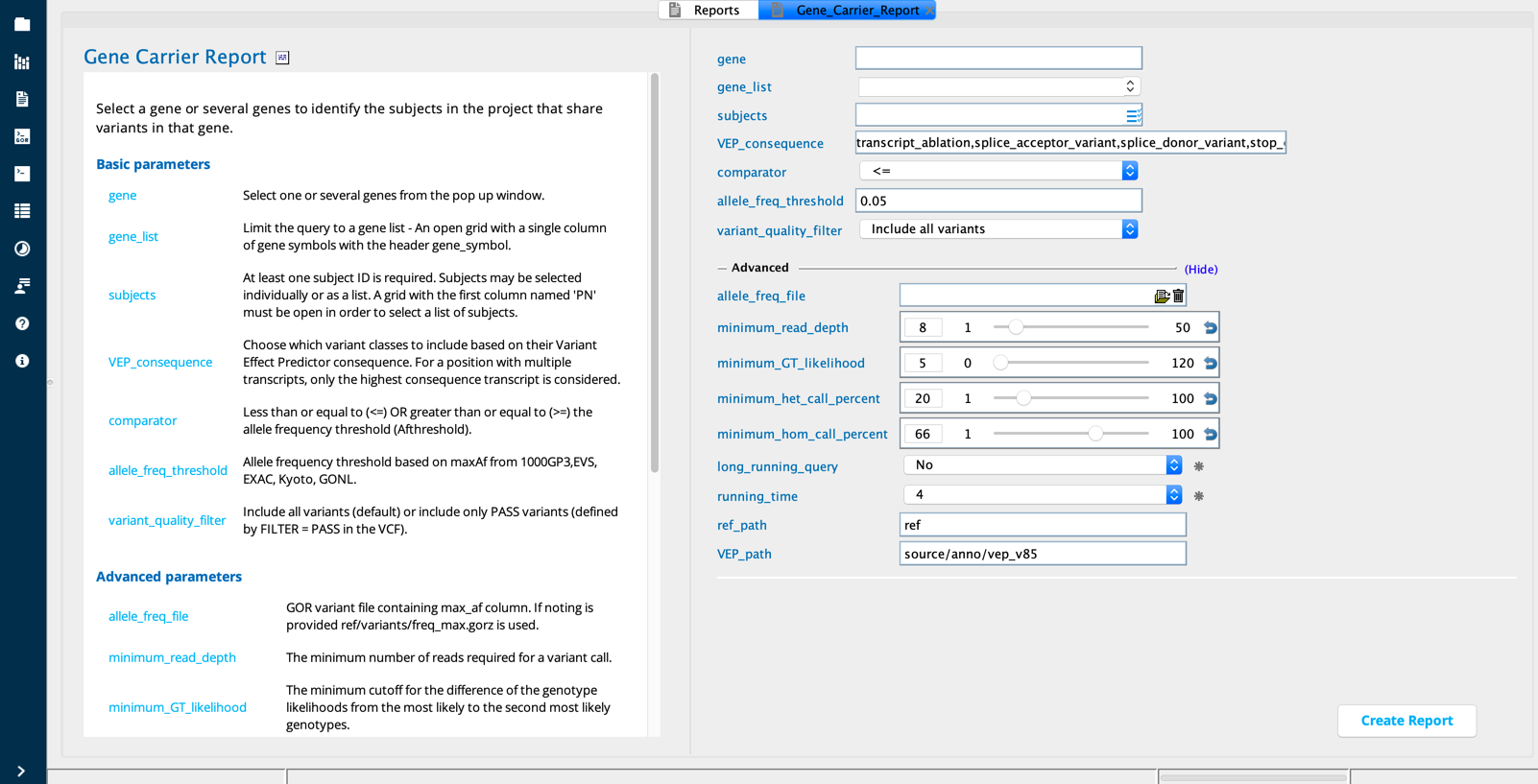
Gene Carrier Report module in Sequence Miner¶
Example Use Case¶
An investigator has a list of candidate genes for autism and wishes to identify all carriers among a subset of patients (samples in the project) of variants with HIGH and MODERATE VEP_consequence in the genelist. The investigator creates a grid of the subject PNs of interest (the SUBJECTS) and a grid of the genelist of interest, sets the desired VEP_consequence threshold, and creates the report.
Description of the algorithm¶
All variants in the user-designated genes/genelist that meet the desired thresholds for VEP_max_consequence, allele frequency, and quality filters are reported in the output with a total count of the carriers for each reported variant.
Interpreting the output¶
This report returns a PN_count column and a lis_PN column. The PN_count column displays the number of samples carrying the variant in that row. The lis_PN column displays a comma-delimited list of the samples (PNs) carrying the variant in that row.
Column descriptions¶
Group |
Column |
Description |
|---|---|---|
Basic |
Call |
|
Chrom |
||
Pos |
||
Reference |
||
Max |
AF |
Maximum allele frequency from public databases (1000Genomes, Exome Variant server, etc.) |
consequence |
Consequence type reported for this variant having the greatest impact |
|
Impact |
Classification of the level of severity of the transcript consequence type assigned by VEP |
|
Other columns |
Amino_Acids |
The amino acid with and without variant (provided only if the variation affects the protein-coding sequence), otherwise “.” |
Biotype |
Biological class of transcript or regulatory feature |
|
CDS_position |
Position of the base pair in the coding sequence; a value is given for each transcript |
|
Gene_Symbol |
Based on HGNC when it exists, otherwise it is the Ensembl internal alias |
|
lis_PN |
A comma-delimited list of the samples containing this variant |
|
PN_count |
The number of samples containing this variant |
|
Protein_Position |
Position of the amino acid in the protein sequence (only if the variant falls within a coding sequence); a value is given for each corresponding transcript specified in the CDS position field |
|
Refgene |
The accession number from NCBI of the affected transcripts |
|
Transcript_count |
Number of different transcripts in which the variant is found |