Sequence Kernel Association Test (SKAT)¶
The Sequence Kernel Association Test (SKAT) is a variance-component test for genetic association analysis. SKAT evaluates the distribution of genetic effects of a group of variants on continuous or dichotomous traits. SKAT detects variants with both positive and negative effects.
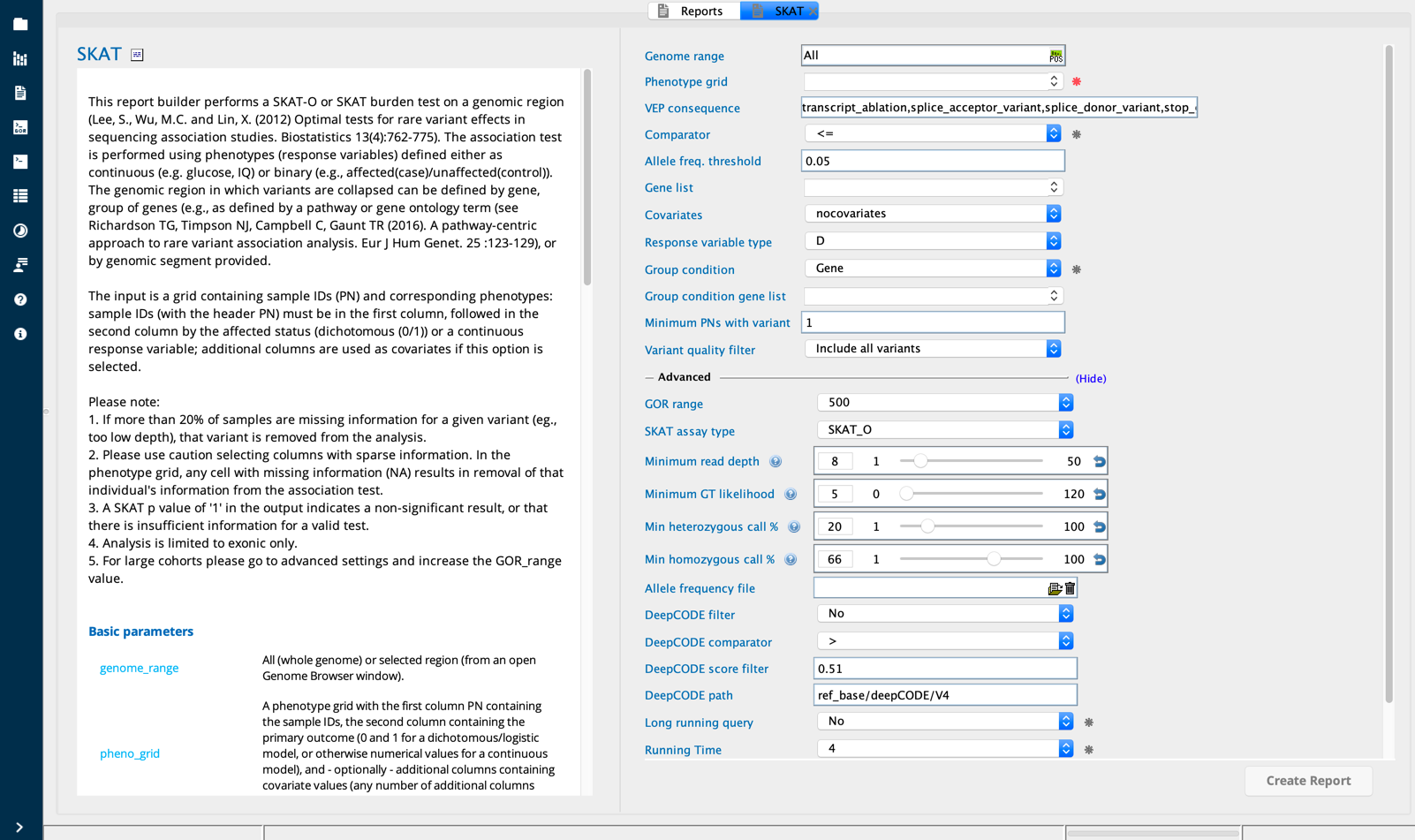
SKAT module in Sequence Miner¶
Example use case¶
The user has genome-wide variant data on cases with hypercholesterolemia and unaffected controls and wishes to evaluate each gene based on the combined evidence for association between the trait and all variants carried in the gene across the cohort.
Description of the algorithm¶
Instead of aggregating variants into an aggregate-allele (e.g., the GeneAnalysis_GAVA report builder), SKAT evaluates the distribution of the aggregated score statistics of individual variants involved in continuous or dichotomous traits. SKAT detects variants with both positive and negative effects. The algorithm is described in Lee et al., Am J Hum Genet 2014 Jul 3; 95(1): 5–23 and Am J Hum Genet 2012 Aug 10;91(2):224-237.
The input phenogrid lists the subjects (PNs) in the first column, the outcome parameter in the second columns, and phenotype scores (numeric values for response variables) in the following columns. The variant genotype data for each subject is extracted from the variant files and the association analysis is performed for each variant genotype vs. phenotype score.
Interpreting the output¶
Genes with p-values less than the user’s threshold for statistical significance (e.g., p-value < 0.05) are considered to exhibit association with the affected status.
Column descriptions¶
Column |
Description |
|---|---|
Chrom |
The chromosome of the variant represented as chr1, chr2, …, chr22, chrXY, chrX, chrY, chrM |
gene_end |
The end base pair position of the gene |
gene_start |
The start base pair position of the gene (zero based, i.e., the position of the base-pair before the first base pair in the gene) |
Gene_Symbol |
Based on HGNC when it exists, otherwise it is the Ensembl internal alias |
skat_pValue |
nominal p-value |