Recessive analysis¶
The Recessive analysis report counts the number of heterozygous, homozygous, and compound heterozygous variants in cases and controls for variants that pass filtering parameters. The report provides a count for each genotype in cases and controls, comprehensive annotation and includes two focused perspectives of the findings:
ARhomAndCHZ– displays only homozygous and compound heterozygous variants that are present in cases and not in controls;AutoRecessHom– displays only homozygous variants that are present in cases and not in controls.
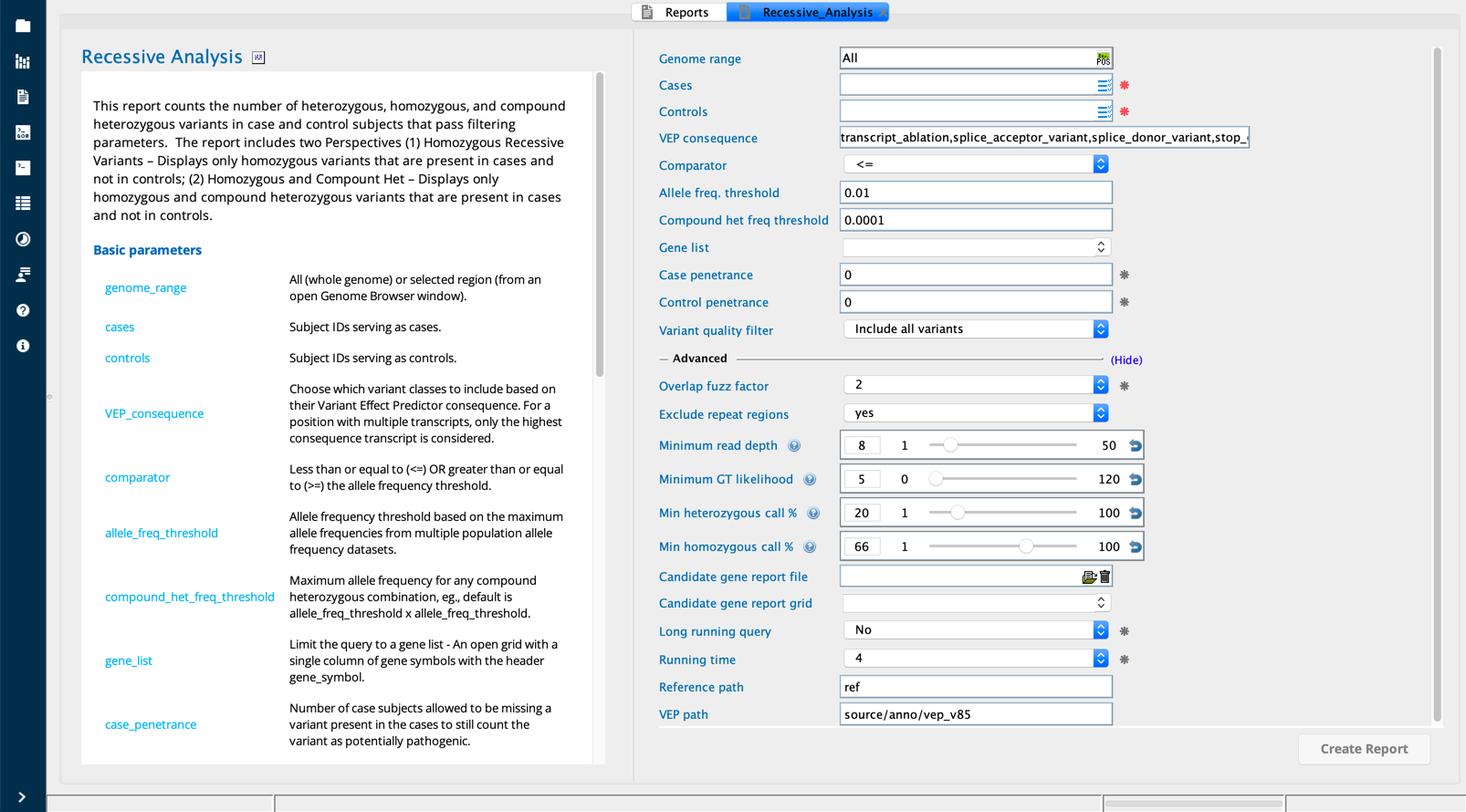
Recessive Analysis module in Sequence Miner¶
Example use case¶
When performing a cohort analysis, the user wishes to compare the distribution of heterozygous versus homozygous calls for particular variants across cases and controls. This report builder calculates the number of controls/cases that are hom/het/chz for each identified variant and adds clinical annotation to all reported variants. These annotation features may be used to prioritize or filter the variants.
Description of the algorithm¶
Variants meeting the user’s criteria for maximum allele frequency and VEP consequence are extracted from whole exome data from the source/anno/vep_v3-4-2/vep_single_wes.gord file and annotated with MaxClinImpact and other clinical information from the ref/clinical_variants.gorz file.
Annotated variants are then filtered to generate two sets:
exome variants set: all variants that reside in exons
exome variants not homozygous in controls set: all variants that meet all three of the following:
Reside in exons
Meet the user’s designated quality criteria for GL_Call, Depth, CallCopies and CallRatio
Are not homozygous in any controls
Each of the variants sets above are then annotated with CAT scores and recessive CAT scores for compound heterozygotes:
STEP 1: CAT scores are calculated as follows:
If the AF > the user’s designated
maxAF threshold(eg: max_AF>0.01), then the variant is CAT4.If the variant is not CAT4 and if
MaxClinImpactis “pathogenic” (in theref/clinical_variants.gorzfile), then the variant is CAT1.If the variant is neither CAT4 or CAT1 and the max_impact is “HIGH” (from the
source/anno/vep_v3-4-2/vep_single_wes.gordfile), then the variant is CAT2.- If the variant is neither CAT1, 2 or 4 and
if
max_Consequence= ‘missense_variant’ andmax_score>= 0.9, the variant is CAT3A, orif
max_impact= ‘LOW’ or (max_impact= ‘MODERATE’ and Cat3A = 0), the variant is CAT3B
STEP 2: An aggregate DIAG_recessiveCAT score is calculated for homozygous and all possible compound heterozygous variant combinations per gene by applying the following seven tests:
2 CAT1 variants or a CAT1 variant in combination with a CAT2 variant,’1:DxConsistent’
either a CAT1 or CAT2 variant in combination with a CAT3A variant,’2:DxLikely’
either 2 CAT3A variants or a CAT1, CAT2 or CAT3A variant in combination with a CAT3B variant , ‘3:DxPossible’
2 CAT3B variants,’4:DxLessLikely’
A CAT1 or CAT2 variant in combination with a CAT4 variants, ‘5:DxIndeterminate’
A CAT3A or a CAT3B variant in combination with a CAT4 variant, ‘6:DxUnlikely’
Variant combinations that don’t meet any of the above criteria,’7:DxNegative’
Heterozygotes, homozygotes, and compound heterozygotes are tallied for cases and controls for each variant that meets the user designated criteria. In addition, DIAG_ACMGcat, DIAG_recessiveCAT, and other annotations are returned for each variant.
Interpreting the output¶
This report builder lists one variant per row. The results fall into three categories: cases, controls, and annotation.
Column descriptions¶
Group |
Column |
Description |
|---|---|---|
Basic |
Call |
|
Chrom |
||
POS |
||
Reference |
||
cases |
het |
The number of cases that are heterozygous for the variant allele |
hom |
The number of cases that are homozygous for the variant allele |
|
WithTheseCHZ |
The number of cases that are compound heterozygous for this variant in this gene |
|
CDG |
AGE_GROUP |
Pediatric: less than 18 years of age; Adult: at least 18 years of age |
CONDITION |
Conditions resulting from mutations in the same gene but may otherwise be placed in the “General” Intervention category |
|
INHERITANCE |
Pattern of inheritance; AD: autosomal dominant; AR: autosomal recessive; BG: blood group; Digenic: a condition resulting from simultaneous mutations in two different genes; Maternal: maternal mitochondrial inheritance; XL: X-linked (as X-linked conditions can frequently have manifestations in both genetic sexes, X-linked conditions are not designated as dominant or recessive) |
|
ctrls |
het |
The number of controls that are heterozygous for the variant allele |
hom |
The number of controls that are homozygous for the variant allele |
|
WithSomeCHZ |
The number of controls are compound heterozygous for this variant in this gene |
|
DIAG |
ACMGcat |
Categorization of the sequence variants according to the ACMG scheme |
recessiveCat |
The recessive and compound heterozygosity category is based on the ACMG category for two alleles, i.e., for any two variants observed in a gene or for a homozygous variant; the ACMG category indicates the likelihood that the variant combination is causative |
|
KNOWN |
Gene_diseases |
Diseases known to be associated with the gene |
var_diseases |
Diseases known to be associated with the variant |
|
OMIM |
Descriptions |
OMIM disease descriptions for the gene |
IDs |
The OMIM ID of the gene |
|
VEP |
max_consequence |
Consequence type reported for the variant having the greatest impact |
Max_Impact |
Classification of the level of severity of the transcript consequence type of the sequence ontology description |
|
Refgene |
The accession number from NCBI of the affected transcripts |
|
Other columns |
Gene_Symbol |
Perspective views¶
The Default view perspective lists all variants shared by all cases. The Perspectives subtabs focus on subsets of columns from the default view.
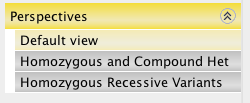
Perspectives in the Gene analysis report builder¶
Perspective |
Description |
|---|---|
Default view |
Lists all variants shared by all cases. |
ARhomAndCHZ |
Lists all variants where the number of cases containing this variant as a compound heterozygote (CHZ) is larger than the total number of controls that carry this variant as a compound heterozygote (CHZ). |
AutoRecessHom |
Lists all variants where the number of cases containing this variant as homozygous is larger than the total number of controls who carry it as homozygous minus the number of CaseDelta. Also the number of controls containing this variant as homozygous is smaller than the CtrlDelta. |