Gene count¶
The Gene count report provides a summary of variants per gene, broken down by VEP consequence or impact, across a set of cases versus controls. For each gene, the number of variants and the number of homozygous variants are tallied for each VEP consequence/impact. The sum total of variants and hom variants, across all VEP consequence/impact categories, are also reported for each gene.
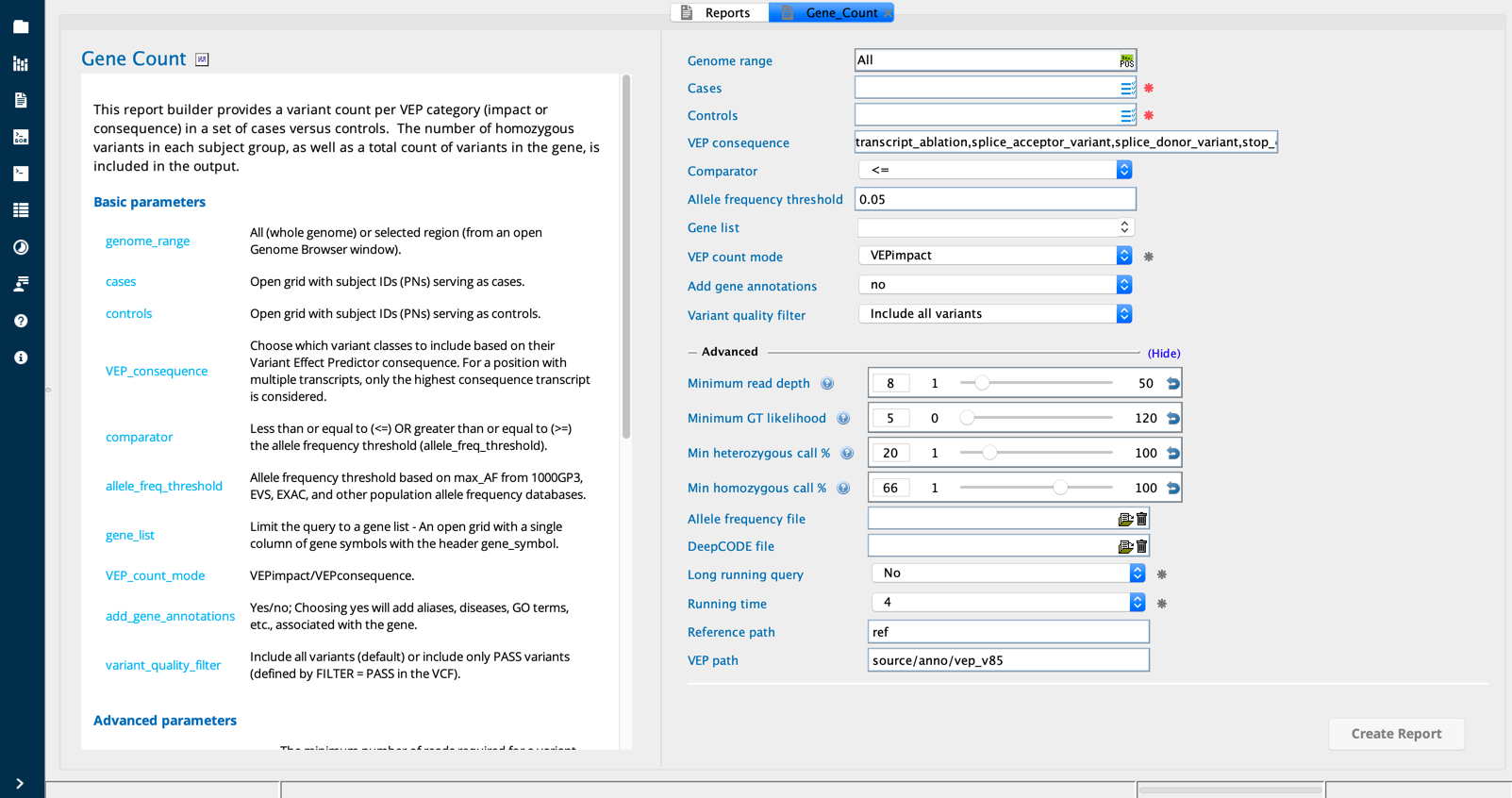
Gene Count module in Sequence Miner¶
Example use case¶
The user wishes to summarize the distribution of variants of several VEP consequence categories across whole exome data in cases versus controls. They also want to evaluate each VEP consequence category distribution per gene individually and then in combination (e.g., frameshift and splice_donor_variant).
Description of the algorithm¶
This query maps filtered variants from the case and control groups onto genes. The filtered variants are then annotated and grouped according to the VEP maximum observed consequence/impact for each gene. The report returns the following for each gene, and for cases and controls separately:
The number of variants within each VEP consequence/impact category
The number of homozygous variants within each VEP consequence/impact category
The sum VEP_Max_Score of variants within each VEP consequence/impact category
The number of variants across all VEP consequence/impact categories
The number of homozygous variants across all VEP consequence/impact categories
The sum VEP_Max_Score of variants across all VEP consequence/impact categories
Interpreting the output¶
The resulting table reports a single row for each VEP impact/consequence (depending on the user selection) per gene. For each category per gene, the following counts are generated:
Variant count (including heterozygous and homozygous) per VEP category per gene per subject group (case or control)
Homozygous variant count per VEP category per gene per subject group (case or control)
Variant count (including heterozygous and homozygous) per gene per subject group (case or control)
Homozygous variant count per gene per subject group (case or control)
Sum of the VEP impact score across a user-designated VEP category per gene per subject group (case or control) (a value between 0 and 1 calculated for each variant as the maximum of (1-SIFT) or Polyphen2 score)
Sum of all of the VEP impact scores across all user-designated VEP categories per gene per subject group (case or control) (a value between 0 and 1 calculated for each variant as the maximum of (1-SIFT) or Polyphen2 score)
Column descriptions¶
Group |
Column |
Description |
|---|---|---|
CASEs |
GENE_homCount |
For each gene, the total number of homozygous variants across all the cases (sum of the CASEs_homCount value in each VEP category for a given gene) |
GENE_sum_Score |
For each gene, the sum of the VEP_max_scores for all the VEP categories across all the cases (sum of the CASEs_sum_Score for each VEP category for a given gene) |
|
GENE_varCount |
For each gene, the total number of variants across all the cases (sum of the CASEs_varCount value in each VEP category for a given gene) |
|
homCount |
The total number of homozygous variants in the VEP category for the given gene across all the cases |
|
sum_score |
The sum of the VEP_max_score of the variants in the indicated VEP category for the given gene across all the cases |
|
varCount |
The total number of variants in the indicated VEP category for a given gene across all the cases |
|
CTRLs |
GENE_homCount |
For each gene, the total number of homozygous variants across all the controls (sum of the CTRLs_homCount value in each VEP category for a given gene) |
GENE_sum_Score |
For each gene, the sum of the VEP_max_scores for all the VEP categories across all the controls (sum of the CTRLs_sum_Score for each VEP category for a given gene) |
|
GENE_varCount |
For each gene, the total number of variants across all the controls (sum of the CTRLs_varCount value in each VEP category for a given gene) |
|
homCount |
The total number of homozygous variants in the VEP category for the given gene across all the controls |
|
sum_score |
The sum of the VEP_max_score of the variants in the indicated VEP category for the given gene across all the controls |
|
varCount |
The total number of variants in the indicated VEP category for a given gene across all the controls |
|
gene |
end |
|
start |
||
Symbol |
||
Other columns |
Chrom |
|
VEP_Max_Impact |
Classification of the level of severity of the transcript consequence type assigned by VEP |
|
VEP_Max_consequence |
VEP predicted consequence for a variant producing the the greatest impact on the transcript (shown only if “VEP_Consequence” is selected in the VEP_count_mode field) |
Drill-in reports¶
Drill-in |
Description |
|---|---|
CaseAndControl_VarsInGene |
For the selected gene variant in the Mendel report, this drill-in report lists all case and controls variants in the same gene with any of the user-selected maximum consequences and the variant genotype for each carrier. |