Compare GOR reports¶
This query compares variants in two variant reports. The output is a table with variant coordinates and a calculated column categorizing variants as follows:
Present in only report 1 (R1)
Present in only report 2 (R2)
Present in both reports (R1,R2)
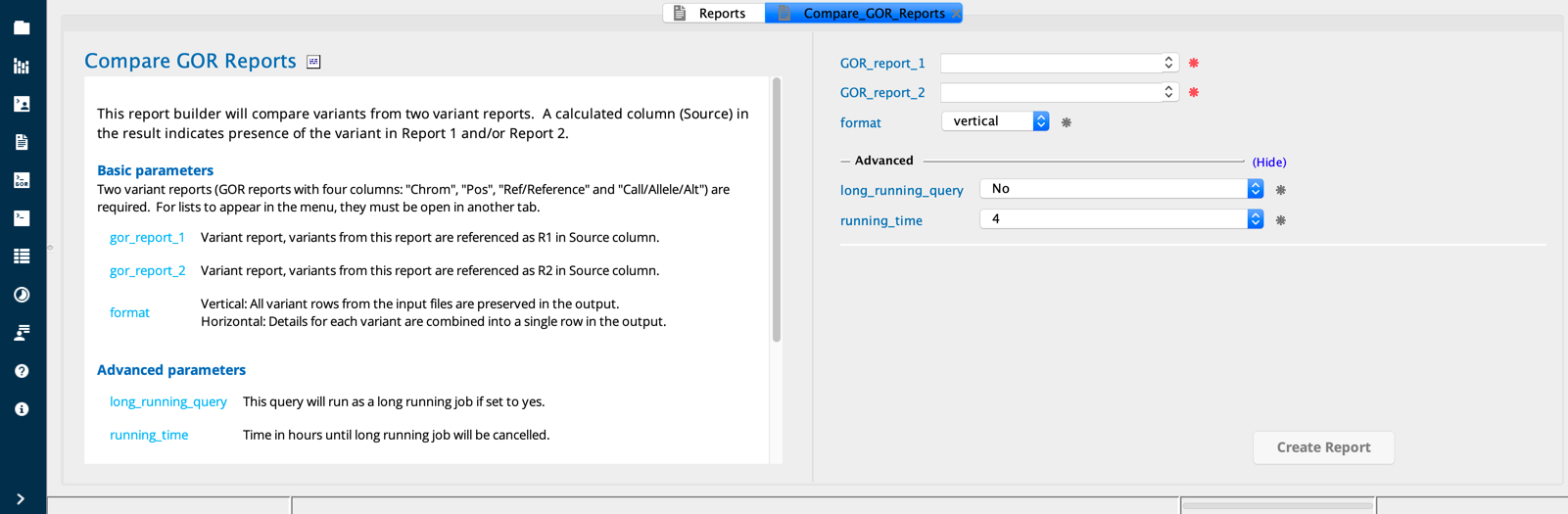
Compare GOR Reports module in Sequence Miner¶
Example use case¶
A user wishes to identify variants common to two possibly genetically-related phenotypes and runs the Multi-family Mendelian analysis report builder separately on two sets of cases:
Cases with progressive vision loss
Cases with progressive hearing loss
The user can run this report builder using each Multi-family Mendelian analysis report as input to identify variants common to both sets of cases and those unique to one set or the other.
Description of the algorithm¶
The algorithm creates a table listing the union of all variants in both reports (all variants in R1 and R2, one variant per row), and calculates a Source column in which each variant is denoted as belonging to the intersection of both reports (R1,R2) or belonging only to R1 or R2.
Interpreting the output¶
The user can choose to have the default display of the output in vertical or horizontal format:
In the vertical format, there is a single PN column listing the index for the report in which the variant is present.
In the horizontal format, each variant has an R1_PN column and an R2_PN column. Only one of these cells can be populated per row. Therefore, if a variant is present in both R1 and R2, then the variant is listed in two rows.
Column descriptions¶
Group |
Column |
Description |
|---|---|---|
Basic |
Call |
The actual called sequence (variant), found by replacing a part of the reference sequence, denoted by Pos and Reference, with the sequence in the Call column |
Chrom |
The chromosome of the variant, represented as chr1, chr2, …, chr22, chrXY, chrX, chrY, chrM |
|
PN |
The patient number (identifier) |
|
Pos |
The (first) base pair position of the sequence variant, e.g., the position of the first nucleotide in the Reference column |
|
Reference |
Sequence from the reference build, the first base starting at the base pair position in the Pos column |
|
Other columns |
CallCopies |
Because the focus is only on variations from the reference, CallCopies refers to how many copies of the variation exist in the subject; “2” corresponds to a homozygous variation whereas “1” corresponds to a heterozygous variation |
CallRatio |
Proportion of reads containing the variation call; expected to be close to 0.5 for heterozygous calls and close to 1 for homozygous calls |
|
Depth |
The number of reads used in evaluating the corresponding call |
|
FILTER |
Quality parameter using the ratio between gt-quality and depth showing if the call is considered of “LowQual” quality (not useable) or “PASS”; this is still a very crude quality measure |
|
formatZip |
VCF genotype fields |
|
FS |
Fisher’s exact test of read strand; if the reference reads are balanced between forward and reverse strands, then the alternate reads should be as well |
|
GL_Call |
A statistical measure indicating the likelihood the call is wrong; the scale has been converted to use only integer numbers - the higher the number, the less likely it is that the call is wrong |
|
Source |
Displays each source report ID; if multiple subjects or reports contain a variant, then the variant will be listed in multiple rows with one report ID per row (e.g., “R1” or “R2”) |
|
Sources |
Displays the variant reports that contain this variant; if multiple reports contain this variant, the value will be a comma-delimited list (“R1,R2,…”) |
Note
Annotation columns are derived from input files.
Perspective views¶
The Default view perspective shows all variants in either report with complete column annotations from source reports. Additional Perspectives focus on subsets of the columns in the default view.
Perspective |
Description |
|---|---|
Both |
Variants in both reports with five columns: Chrom, POS, Reference, Call, Sources |
Default view |
Shows all variants in either report with complete column annotations from source reports |
Either |
Variants present in either report with five columns: Chrom, POS, Reference, Call, Sources |
R1only |
Variants uniquely found in R2 report with five columns: Chrom, POS, Reference, Call, Sources |
R2only |
Variants uniquely found in R2 report with five columns: Chrom, POS, Reference, Call, Sources |